Epidemiological Analysis of Porcine Circovirus Type 2 (PCV2) Spatio-temporally of Circulation in Vietnam from 2004 to 2019
- School of Biotechnology, Tan Tao University, Tan Duc E.City, Duc Hoa, Long An, 850000, Viet Nam
- School of Biotechnology, Tan Tao University, Tan Duc E.City, Duc Hoa, Long An, 850000, Vietnam.
- Faculty of Animal Science and Veterinary Medicine, Nonglam University HCMC, Thu Duc district, Ho Chi Minh City, Vietnam
- bFaculty of Animal Science and Veterinary Medicine, Nonglam University HCMC, Thu Duc district, Ho Chi Minh City, Vietnam
- Research Center for Genetics and Reproductive Health, School of Medicine, Vietnam National University Ho Chi Minh City, Linh Trung ward, Thu Duc district, Ho Chi Minh City, Vietnam
- Department of Biomedical Engineering, School of Medicine, Vietnam National University Ho Chi Minh City (VNU-HCM), Ho Chi Minh City 700000, Vietnam
- Vietnam National University Ho Chi Minh City (VNU-HCM), Ho Chi Minh City 700000, Vietnam.
Abstract
Introduction: Porcine circovirus 2 (PCV2) is a causative agent of postweaning multisystem wasting syndrome (PMWS), an epizootic disease that causes significant economic losses to pig producers worldwide, including Viet Nam. However, analyses of which groups of PCV2 strains cause disease in pigs in Viet Nam are still limited, leading to the delay or lack of application of specific vaccine sources to prevent PMWSs in other countries.
Method: In this study, we aimed to analyze the geographical and temporal diversity of PCV2 in pig farms across 26 provinces in Vietnam, thus identifying the dominant strain for vaccine production. After samples from North, Central, and South Viet Nam were collected, phylogenetic trees were created to study the molecular genetic/evolutionary relationships, thereby classifying the existing PCV2 strains. The spatiotemporal distribution of strains is shown visually for observation. Finally, an analysis of the important evolutionary differences between the two strains, PCV2d and PCV2b, revealed that these strains were superior in Viet Nam.
Results: Our study examined the diversity of PCV2 strains in Vietnam, ranging from the northern to southern regions, and the PCV2d strain was revealed to be the predominant strain. Interestingly, we observed the presence of 2 or 3 different genotypes of PCV2 in the same province and at the same time. We identified the presence of PCV2h, PCV2g, and PCV2e genotypes that had not been previously detected. Furthermore, we identified potential sites of positive selection in the capsid epitopes, which are believed to play a crucial role in evading the host immune system.
Conclusions: The results showed that Viet Nam has 5 genotypes, namely, PCV2b, PCV2e, PCV2g, PCV2h and PCV2d—the main causative strains of the disease. The coexistence of strains in the same geographical area was also shown, indicating the importance of preparing vaccines and preventing livestock infection in Viet Nam.
INTRODUCTION
Postweaning multisystem wasting syndrome (PMWS) in swine was first discovered in 1991 in Canada and is characterized by emaciation and jaundice 1, 2. Over the years, common disease manifestations have been described as emaciation, pale skin, jaundice, and diarrhea in piglets 3. After years of research and sequence analysis, PMWS has been found to be caused by porcine circovirus (PCV)4, 5. PCV type 1 (PCV1) is a virus derived from cell culture, whereas PCV type 2 (PCV2) is a virus strain associated with swine diseases6. PCV2 causes a disease collectively known as porcine circovirus-associated disease (PCVAD or PMWS), which, although emerging globally, has a major impact on the global swine industry, causing significant economic losses to many specialized pork export countries, including Viet Nam. Subsequent studies have shown the relationship of PCV2 with reproductive disorders7, 8, enteritis 9, 10, respiratory disease11, 12, dermatitis, and other syndromes, such as porcine dermatitis and nephropathy syndrome (PDNS)13, 14 and proliferative and necrotizing pneumonia (PNP)3, 10.
Pigs of all ages are susceptible to PCV2, and symptoms of PMWS are commonly observed between 6 and 10 weeks of age15. Mortality rates in pigs can range from 4 to 20% 16. PCV2 can be found in various secretions (eyes, nose, bronchi), saliva, urine, milk, and semen, indicating diverse transmission routes both horizontally and vertically17, 18, 19, 20, 21, 22. Horizontal transmission occurs primarily through the nose and mouth23. Opriessing et al. confirmed transmission through the pig's mouth by feeding uncooked food from infected pigs24. The most common vertical transmission route occurs through the placenta from mother to offspring, leading to fatal PCV2 infection22.
PCV1 and PCV2 belong to the Circoviridae family10, 25, 26 and the genus Circovirus27. This family has a specific avian host and a relatively narrow host range. These viruses are small, nonenveloped, icosahedral viruses with a single-stranded DNA genome (1767/1768 bp) that includes 7 open reading frames (ORFs). Among these, ORF1 encodes a replicase (Rep) protein, and ORF2 encodes a capsid (Cap) protein. Rep and Cap are important components of infectious virions 28, 4. The PCV2 genome has a conserved stem loop structure in its ssDNA, which allows it to infect eukaryotes29. The rate of nucleotide substitution for PCV2 was estimated to be on the order of 1.2 × 10 substitutions/site/year, the highest ever observed for a ssDNA virus 30, 31, 32. This high evolutionary rate could facilitate the rapid emergence of PCV2 worldwide.
Because ORF2 has a higher evolution rate than the whole genome of PCV2 and is under great selective pressure from the immune system33, PCV2 can enter host cells. On the other hand, the ORF2 cap is considered a unique protein structure that plays a role in determining the antigenicity and virulence of PCV2 4. Therefore, changing the amino acid sequence of ORF2 can change its pathogenicity and virulence. For these reasons, ORF2 has become the focus of many studies.
The convention to genotype PCV2 based on the prevalent diversity of the ORF2 nucleotide was accepted in 200834. Studies of the PCV2 genome have identified 8 genotypes, namely, PCV2a, PCV2b, PCV2c, PCV2d, PCV2e, PCV2f, PCV2g, and PCV2h31, 32, 35, 36, 37, 38. Genotype shifts may be associated with differences in pathogenicity and vaccine immunity39. These remarkable "genotype shifts" revealed that PCV2b displaces PCV2a as the dominant virus and has increased virulence31, 40, 41, 42. On the other hand, the mutant PCV2b-calling PCV2d strains isolated from PCV2b strains have begun to gain increasing attention for pathogenicity in many countries, such as China, Korea, the United States, and South America43, 44. Therefore, there is a need to identify novel PCV2 genotypes to develop more effective vaccines for rapidly evolving PCV2.
In Viet Nam, studies on new strains of PCV2 are limited. This study aimed to investigate the annual geographical distribution of PCV2 across Viet Nam. The results of this study will provide an objective, accurate assessment of the current raging strains of PCV2, as well as a direction for successful vaccine development.
MATERIALS AND METHODS
Sample collection, DNA isolation, PCR, and sequencing
From 76 pig farms, samples were collected from 127 pigs with clinical PRDC manifestations (Figure 1). The tissue samples were processed and frozen (-20°C) within 12 hours. Three grams of sample was homogenized in a 15 ml Falcon tube by adding PBS to a 10% suspension. The tissue homogenate was subsequently centrifuged at 3000 rpm for 3-5 min at 4°C. One to three millilitres of the supernatant was used for nucleic acid isolation. Viral nucleic acid was extracted from tissues using the GeneJET Viral DNA/RNA Purification Kit (Thermo Scientific, USA) according to the manufacturer’s instructions. PCR was used to identify potential cases of infection with PCV2 using the primers PCV2F and PCV2R, which were designed based on 66 ORF2 sequences of the PCV2 strain. PCR was carried out in 25 µl reaction mixtures containing 10 µl of Green Master Mix (GoTaq Green Master Mix, Cat# M7122; Promega, USA), 3 µl of DNA template, 0.5 µl of 10 µM each primer, and 10.5 µl of nuclease–free water. The PCR temperature cycling conditions were as follows: 95°C for 3 min; 33 cycles of denaturation at 94°C for 60 s and 58°C for 60 s; 72°C for 45 s; and 72°C for 5 min. The PCR products were then separated and visualized via 1.2% agarose gel electrophoresis with Gel-red staining. The 702-bp bands of 43 samples (potential PCV2 strain) were collected and purified using the Wizard Preps DNA Purification and PCR Clean-Up System (Promega, USA). The purified products were subsequently sent to Macrogen (Seoul, Korea) for sequencing using the PCV2R and PCV2F primers. These sequences are available in GenBank under accession number MT432425-67. After sequencing, 42/43 ORF2 sequences were selected for phylogenetic study (one sample was disqualified due to ineligibility of the Beast tool).

A total of 76 pig farms in the provinces of Viet Nam were sampled (2018-2019).
PCV2 sequences in Viet Nam were collected from the NCBI
To obtain the most approximate results, it is necessary to have a large amount of information as well as the sequences to be analyzed in all three regions of North-Central-South Viet Nam. Therefore, in this study, two additional sources of sequences from the North were identified to enrich the PCV2 sequence of Viet Nam and help the study obtain more reliable results. First, 43 ORF2 sequences of the PCV2 strain from previous studies were collected from the GenBank. These PCV2 strains were isolated from several provinces of Viet Nam from 2004 to 2017, and their names are listed in
Altogether, a total of 87 ORF2 nucleotide sequences from 26 provinces in the northern, central, and southern regions of Viet Nam from 2004 to 2019 were used for analysis in this study. The genome size of the 87 PCV2 strains collected in this study was 626 nucleotides after alignment and editing of the sequences.
The sequence IDs of the PCV2 strains were obtained from 2004–2019 from 26 provinces in Viet Nam
|
No. |
Strain (1) |
Location (2) |
Year (3) |
No. |
Strain |
Location |
Year |
|---|---|---|---|---|---|---|---|
|
1 |
NAVET_vietnam3 |
BD |
2004 |
44 |
DTD-AnPha/BD |
BD |
2018 |
|
2 |
749-Vp33 |
VP |
2004 |
45 |
DTD-CS1/ND |
2018 | |
|
3 |
P2425NT |
TN |
2008 |
46 |
DTD-Dat/ND |
2018 | |
|
4 |
550-TH |
TH |
2008 |
47 |
DTD-Guyo/DN |
DN |
2018 |
|
5 |
641-HB |
HB |
2008 |
48 |
DTD-Hai/BP |
2018 | |
|
6 |
P473BY |
YB |
2009 |
49 |
DTD-Minh/ND |
2018 | |
|
7 |
P477LG |
GL |
2009 |
50 |
DTD-N/ND |
BTh |
2018 |
|
8 |
P485YH |
HY |
2009 |
51 |
DTD-Nhon/DN |
DN |
2018 |
|
9 |
P624LB |
BL |
2009 |
52 |
DTD-Phuoc2/TG |
TG |
2018 |
|
10 |
661-BN |
2009 |
53 |
DTD-RT2/GL |
GL |
2018 | |
|
11 |
529-Hno |
Han |
2009 |
54 |
DTD-RT5.2/DL |
DL |
2018 |
|
12 |
P789NH |
HN |
2010 |
55 |
DTD-RT5/DL |
DL |
2018 |
|
13 |
P902NH |
HN |
2010 |
56 |
DTD-T/ND |
BTh |
2018 |
|
14 |
638-HY |
HY |
2010 |
57 |
DTD-The/ND |
2018 | |
|
15 |
HD1-9 |
HD |
2011 |
58 |
DTD-Tiep/QN |
QNg |
2018 |
|
16 |
HB1-4 |
HB |
2011 |
59 |
DTD-Tuan/ND |
2018 | |
|
17 |
HB1-6 |
HB |
2011 |
60 |
DTD-Tuyen/LD |
LD |
2018 |
|
18 |
BG0-1 |
BG |
2011 |
61 |
DTD-03PG/BD |
BD |
2019 |
|
19 |
Han13-4 |
Han |
2011 |
62 |
DTD-07PG/BD |
BD |
2019 |
|
20 |
Han6-12 |
Han |
2011 |
63 |
DTD-09PG/BD |
BD |
2019 |
|
21 |
TN1-1 |
TN |
2011 |
64 |
DTD-10PG/BD |
BD |
2019 |
|
22 |
TN4-1 |
TN |
2011 |
65 |
DTD-118/HCM |
HCM |
2019 |
|
23 |
TN4-M1 |
TN |
2011 |
66 |
DTD-11PG/BD |
BD |
2019 |
|
24 |
TN5-3 |
TN |
2011 |
67 |
DTD-125/HCM |
HCM |
2019 |
|
25 |
HD1-1 |
HD |
2011 |
68 |
DTD-C2/DN |
DN |
2019 |
|
26 |
Han0-4 |
Han |
2011 |
69 |
DTD-C3/DN |
DN |
2019 |
|
27 |
HB1-1 |
HB |
2011 |
70 |
DTD-Cao/ND |
2019 | |
|
28 |
Han14 |
Han |
2011 |
71 |
DTD-CH4/BT |
BTh |
2019 |
|
29 |
HB2 |
HB |
2011 |
72 |
DTD-Cong/ND |
2019 | |
|
30 |
178-HnDfu |
Han |
2011 |
73 |
DTD-CS7 W/VP |
VP |
2019 |
|
31 |
002-Hna |
HN |
2011 |
74 |
DTD-HMT/DL |
DL |
2019 |
|
32 |
BG58 |
BG |
2012 |
75 |
DTD-N1/ND |
2019 | |
|
33 |
PT6 |
PT |
2013 |
76 |
DTD-N3/ND |
2019 | |
|
34 |
NN4 |
Han |
2013 |
77 |
DTD-NA4/DN |
DN |
2019 |
|
35 |
M2 |
2014 |
78 |
DTD-NB3/DN |
DN |
2019 | |
|
36 |
Hau7 |
Han |
2014 |
79 |
DTD-NC2/DN |
DN |
2019 |
|
37 |
Hau8 |
Han |
2014 |
80 |
DTD-Phuc/HCM |
HCM |
2019 |
|
38 |
Hau9 |
Han |
2014 |
81 |
DTD-Phuoc1/ND |
2019 | |
|
39 |
Hau2 |
Han |
2014 |
82 |
DTD-SP1/Bdi |
BDi |
2019 |
|
40 |
Hau3 |
Han |
2014 |
83 |
DTD-SP2/Bdi |
BDi |
2019 |
|
41 |
Hau5 |
Han |
2014 |
84 |
DTD-Trong/ND |
2019 | |
|
42 |
M1 |
2014 |
85 |
DTD-Vinh/DN |
DN |
2019 | |
|
43 |
Hau6 |
Han |
2014 |
86 |
PCV2/G22111 |
TB |
2018 |
|
87 |
PCV2/G40312 |
PT |
2018 | ||||
Phylogenetic analysis
The sequences for phylogenetic analysis were divided into four datasets: 42 PCV2 strains from this study (2018-2019), 43 PCV2 strains (2004-2017), 2 PCV2 strains (2018) from a previous study in Viet Nam, and 42 PCV2 reference strains from other countries (Table S1). Details of the 87 PCV2 strains from Viet Nam are provided in
All 129 sequences used in the study were aligned with ClustalW and edited via the Molecular Evolutionary Genetics Analysis (MEGA) X tool (10.2.4). The phylogenetic tree was subsequently constructed with the MEGA X tool based on the ORF2 sequence using neighbor-joining (NJ) trees (classified according to evolutionary distance) and maximum composite likelihood (ML) methods (scores of likelihoods in classification). PCV1 was selected as an outgroup element. The implemented bootstrap value was 1000. With the NJ method, a phylogenetic tree was built based on the Kimura 2-parameter model. Like in the NJ method, in the ML method, the uniformity rate among sites was determined. In addition, the maximum clade credibility (MCC) tree (representing the time scale phylogeny) was also built through BEAST (v1.10.4) software and TreeAnnotator (v1.10.4) and then visualized with FigTree (v1.4.4). The BEAST parameters used were established according to previous methods https://beast.community/first_tutorial. The distribution map of Viet Nam was drawn using ArcMap software 10.8.
Positive selection analysis
After phylogenetic analysis, the sequences of the PCV2b and PCV2d strains were further analyzed via selective pressure analysis. Sequences with a length of 624 nucleotides (Cap protein coding region - ORF2) were uploaded to Data Monkey (www.datamonkey.org), analyzed and selected based on p value calculations 46. To estimate the sites under selection, four algorithms were used, including single-likelihood ancestor counting (SLAC), mixed effects model of evolution (MEME), fixed effects likelihood (FEL), and fast unconstrained Bayesian approximation (FUBAR)46, 47, 48. A position was potentially positively selected if it satisfied one of four algorithms with p < 0.1 in SLAC, p < 0.05 in FEL and MEME, and p > 0.9 in FUBAR; otherwise, it was considered negative selection. Finally, the cap sequences of the strains PCV2b and PCV2d were used to predict the 3D protein structure submitted to TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) and visualized with PyMOL software (South San Francisco, CA, USA).
RESULTS
In this study, 3 reliable tree generation methods, namely, NJ, ML, and MCC, were used to construct a phylogenetic tree based on 129 ORF2 sequences of PCV2. Using the NJ and ML tree methods, the data showed that PCV2 strains collected from pig farms in Viet Nam exhibited various genotypes (PCV2b, PCV2d, PCV2e, PCV2h and PCV2g) (Figure 2. A and B). None of the PCV2 strains were clustered into PCV2a, PCV2c, or PCV2f. The most prevalent PCV2 strain was PCV2d (54.02%), the second was PCV2h (25.29%), and the third was PCV2b (16.09%). The phylogenetic tree constructed by the MCC method also produced similar results (Figure 3). Interestingly, the results of classifying the sequences on the phylogenetic tree were the same despite the use of different algorithms to generate the phylogenetic tree.

(A) An NJ phylogenetic tree was built with the MEGA X tool using 87 ORF2 PCV2 sequences. PCV2 strains were identified based on reference sequences. The implemented bootstrap value is 1000; (B) An ML phylogenetic tree was built with the MEGA X tool using 87 ORF2 PCV2 sequences. PCV2 strains were identified based on reference sequences. The implemented bootstrap value is 1000.

MCC tree constructed using BEAST (v1.8.4) from 87 Vietnamese PCV2 isolates. The PCV2 sequences were included for classification over time. The posterior region was displayed along each branch. The different strains are represented by different colors, as displayed in the figures. The scale bar represents time (in years).
To determine the optimal use of these vaccines based on the distribution of PCV2 strains in Viet Nam in each province at a given time, we analyzed 87 ORF2 sequences of the PCV2 strains collected in Viet Nam from 2004 to 2019 spanning more than 26 provinces (Figure 4 and

The spatial distribution of PCV2 strains (province) in Viet Nam. The different strains are represented by different colors, as displayed in the figure.
Summary of the spatial genotype information for 87 ORF2 PCV2 sequences subjected to epidemiological analysis (the sequences are colored North, Central, and South) over time
|
Year/Location |
2004 |
2008 |
2009 |
2010 |
2011 |
2012 |
2013 |
2014 |
2018 |
2019 |
|---|---|---|---|---|---|---|---|---|---|---|
|
YB |
2 h | |||||||||
|
HB |
2 g |
2 h - 2e - 2b | ||||||||
|
TNg |
2 g |
2 h | ||||||||
|
PT |
2 h |
2d | ||||||||
|
BG |
2 h |
2b | ||||||||
|
BN |
2b | |||||||||
|
HN |
2 h |
2 h | ||||||||
|
Han |
2e |
2 h - 2b |
2 h |
2d | ||||||
|
HD |
2 h - 2b | |||||||||
|
HY |
2 h |
2 h | ||||||||
|
NB |
2b-2d | |||||||||
|
TB |
2d | |||||||||
|
VP |
2 h |
2b | ||||||||
|
TH |
2 h | |||||||||
|
QNg |
2d | |||||||||
|
BDi |
2d | |||||||||
|
BTh |
2d |
2d | ||||||||
|
GL |
2 h |
2d | ||||||||
|
DL |
2d |
2d | ||||||||
|
LD |
2d | |||||||||
|
BP |
2b | |||||||||
|
BD |
2 h |
2d |
2d | |||||||
|
DN |
2d |
2b-2d | ||||||||
|
HCM |
2d | |||||||||
|
TG |
2b | |||||||||
|
BL |
2 h |
In general, our phylogenetic analysis showed that PCV2d, PCV2h and PCV2b were the most prevalent strains from 2004 to 2019. However, after 2013, PCV2h was no longer present in the livestock samples (
Selection analysis of PCV2 ORF2 (cap) sequences from PCV2b and PCV2d
|
Site |
FEL (0 positive site) |
(2 positive sites) MEME |
(0 positive site) SLAC |
(1 positive site) FUBAR | ||||
|---|---|---|---|---|---|---|---|---|
|
dN-dS (a=b) |
P<0.05 |
W+ (b+) |
P<0.05 |
dN-dS |
P<0.1 |
dN-dS (b-a) |
Post.pro>0.9 (pro a<b) | |
|
59 |
1.619 |
0.98 |
87.52 |
0.02 |
0.131 |
0.448 |
0.153 |
0.563 |
|
63 |
1.955 |
0.22 |
2.49 |
0.24 |
6.78 |
1.00 |
5.092 |
0.908 |
|
68 |
1.736 |
0.665 |
18.86 |
0.04 |
2.54 |
0.62 |
2.594 |
0.751 |
DISCUSSION
Phylogenetic tree analyses (NJ, ML, and MCC) were conducted based on the obtained ORF2 sequences. The PCV2 strains identified in Viet Nam included PCV2b, PCV2d, PCV2e, PCV2h, and PCV2g. PCV2d was the most prevalent strain, consistent with the findings of previous studies49, 50. It appears that PCV2d is increasingly dominant in causing disease in Vietnamese individuals and worldwide swine51, consistent with what was shown in this study. Similarly, in another study in Viet Nam, Nguyen . collected samples and clearly identified 8/13 as the PCV2d strain in southern Viet Nam52. The second most common genotype was PCV2h (25.29%), and the third was PCV2b (16.09%). However, the PCV2h genotype was not considered and was included in the follow-up analysis (positive model selection) because it ceased to occur after 2013 based on the results of the phylogenetic tree analysis and the records. Therefore, the genotypes PCV2b and PCV2d were the focus of this study. In comparison, with the findings of previous studies, three new strains were discovered in our study (i.e., PCV2h, PCV2g, and PCV2e), which were previously identified as PCV2b and PCV2d strains52, 50, 49. However, further studies with larger sample sizes are needed to confirm these findings, considering the influence of the analysis method and reference strains on the overall conclusion.
PCV2 has been recognized as the primary cause of swine diseases worldwide since the late 1990s32. Genotypes such as PCV2a, PCV2b, and PCV2c were classified based on nucleotide variations in ORF234. PCV2b, present in Europe and Asia since 199736 and in North America since 2005 40, is one of the known genotypes. PCV2c was identified in Denmark in 1980 36. PCV2d and PCV2e, two new genotypes, were reported in China in 200953. PCV2d likely emerges as a mutation of PCV2b54, 43, 33. Mutant PCV2b (mPCV2b) strains were also detected in North and South America in 201255, 39. Pham reported that PCV2d strains in Viet Nam originated from China50. PCV2d represents a notable example of a genotype substitution, such as when PCV2b evolves into PCV2d, possibly driven by natural selection or vaccine-related factors.
This study investigated the molecular epidemiology of PCV2 strains based on geography and time, highlighting the diversity and dominance of the strains. Geographically, PCV2h, PCV2b, and PCV2d are distributed across provinces in Viet Nam, with PCV2d being the most prevalent. PCV2b and PCV2d strains were recently identified. The next section will focus on analyzing the positive positions of these two genotypes. The cooccurrence of PCV2b and PCV2d in the same locality indicates variability and potential vaccine inefficiencies. Attention should be given to this coinfection for the development of an effective vaccine in Viet Nam. Furthermore, strict management methods are necessary to prevent interlocality infections and reduce the diversity of PCV2 genotypes causing diseases.

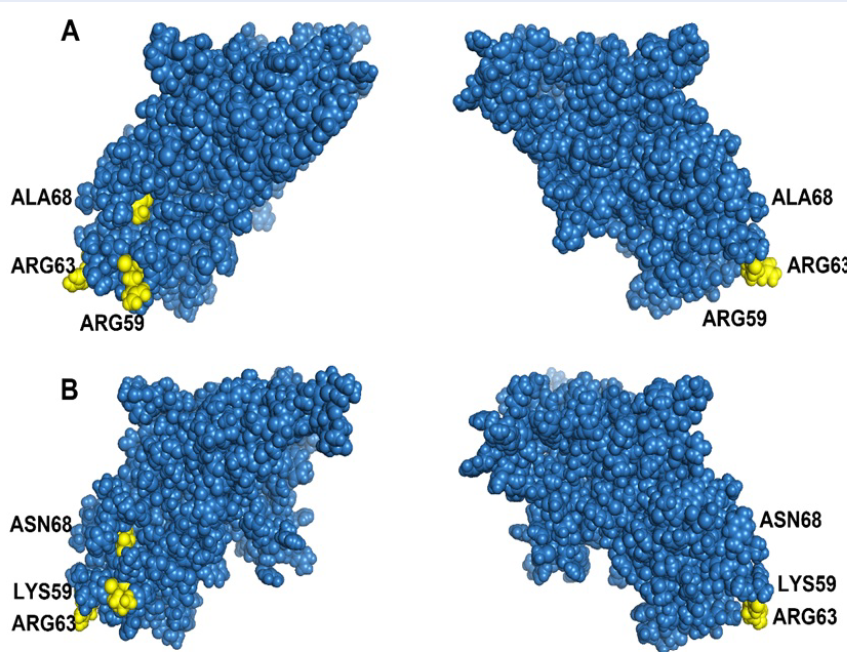
The structure of the PCV2 Cap protein was predicted by using I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). The green filled circles indicate the epitopes of the PCV2 cap protein, and the yellow filled circles indicate the amino acids selected based on positive selection. (A) PCV2b and (B) PCV2d.
Selection analysis revealed that three amino acid sites could be considered potential immune system components, indicating that these sites are markers of positive selection during the specific evolution of PCV2b and PCV2d11, 56, 57, 58, 59, 60. In other words, these amino acid sites could shift PCV2b to PCV2d. These results indicate that the antigenicity of PCV2b differs from that of PCV2d. Codon sites with potential for positive selection are shown on the basis of the conformational prediction of the PCV2 cap protein (Figure 5). These results and phylogenetic analysis might provide insight into the genotypic classification of PCV2.
Selected positions relevant to functional activities, including epitopes in the capsid protein, were identified in previous research. For example, codon 59 is crucial for conformational neutralizing epitopes, and changes in amino acids at this position can strongly affect the immunogenicity of a PCV2 genotype 56. Residue region 165-200 interacts with residues 58-63 to form conformational neutralizing epitopes 57. Codons 63 and 68 are immune-related epitopes58, 59, 60 potentially involved in evading the host immune system. Understanding these positive selections helps predict virulence changes and aids in PCV2 research. The 3D structure of the capsid protein is also important because it reveals the impact of amino acids on virion expression and the environment61. The capsid protein can mutate to evade the host immune system or enhance binding to receptors 62.
Various PCV2 variants have been identified in Viet Nam, posing challenges for the development of an effective PCV2 vaccine. Inactivated vaccines, including those targeting the PCV2a and PCV2b genotypes, have shown promising results in improving pig weight and reducing mortality63, 64. Recombinant vaccines, such as Suvaxyn PCV2® One doseTM, have been developed by integrating PCV2a ORF2 into PCV165. However, the detection of recombinant viruses in pig herds led to the discontinuation of these vaccines, indicating incomplete virus inactivation. Recombinant DNA technology has allowed the production of PCV2 Cap protein-expressing vaccines, such as those based on the PCV2a and PCV2b genotypes66, 67, 68. These vaccines have demonstrated effectiveness in pigs. Currently, imported commercial vaccines circulating in Viet Nam primarily target the PCV2a genotype37. Vaccines such as Cirovac, circo pigvac, and Fostera PCV2 conjugate, which are used in Viet Nam, are also based on the PCV2a genotype69.
Effective vaccines for PCV2 should consider the prevailing strains in Viet Nam, particularly PCV2b and PCV2d 49, 50. Similarities between the vaccine strain and local strains are crucial for vaccine efficacy. The presence of PCV2, especially PCV2d, poses significant risks to Viet Nam's economy and global health. Therefore, the development of dedicated vaccines targeting novel PCV2 subtypes is essential for timely prevention and mitigation of the severe consequences associated with PCV2.
CONCLUSIONS
This study identified diverse PCV2 genotypes in Viet Nam, including PCV2b, PCV2d, PCV2e, PCV2g, and PCV2h. PCV2d was the most common genotype (47/87). The coexistence of multiple genotypes at the same location suggests potential challenges in vaccine effectiveness. Phylogenetic analysis revealed that the PCV2g, PCV2h, and PCV2e genotypes form distinct branches. These findings provide valuable insights into the distribution and diversity of PCV2 strains in Viet Nam. These findings can guide the optimal allocation of vaccine resources and the development of region-specific vaccines in Viet Nam.
ABBREVIATIONS
AA: Amino acid
bp: Base pair
Cap: Capsid
DNA: Deoxyribonucleic acid
MCC: Maximum clade credibility
ML: Maximum likelihood
NJ: Neighbor-joining trees
nt: Nucleotide
ORF: Open reading frame
PCR: Polymerase chain reaction
PCV2: Porcine circovirus type 2
PMWS: Postweaning multisystemic wasting syndrome
PRCD: Porcine respiratory disease complex
Rep: Replicase
RNA: Ribonucleic acid
Author Contributions
MPNN, MNN, and DTD designed the study. MPNN, MNN, and DTD performed the experiments. MPNN, MNN, TTN and THN analyzed the data. MPNN, TNL, and MNN wrote the manuscript. All authors read and approved the final manuscript.
COMPETING INTERESTS
All the authors have read the journal's policy on disclosing potential conflicts of interest, and we declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ACKNOWLEDGEMENTS
We are grateful to the Ho Chi Minh City University of Agriculture and Forestry for enabling us to research and provide technical assistance in sample collection and analysis.

