
Application of anthracnose resistance-associated molecular markers in the detection of resistant chili pepper cultivars in Vietnam
- University of Science, Vietnam National University, Ho Chi Minh city
Abstract
Introduction: Colletotrichum species is responsible for anthracnose, a worldwide serious disease, causing an important loss in chili pepper production. Therefore, screening disease resistant and sensitive chili pepper cultivars in Vietnam is important not only for in-depth studies of disease resistance-associated molecular mechanisms but also for chili production improvement via molecular marker-assisted breeding in Vietnam.
Methods: To this end, in this study, two Colletotrichum isolates were obtained from the infected fruits collected from chili pepper (Capsicum annuum) fields in Tra Vinh province. According to the morphology analysis and the sequencing results of the internal transcribed spacer (ITS) regions, these isolates were identified as C. scovillei and C. acutatum. In order to identify the anthracnose-resistant chili pepper cultivars, the pathogenicity test was conducted by infecting fully developed green fruits of eleven chili pepper cultivars with the two isolated Colletotrichum strains.
Results: CN404 and HNCS were the two strongest anthracnose-resistant cultivars. Two chili pepper cultivars, TV3 and PN400, showed different resistance tendencies to each Colletotrichum isolates. Four different SSR molecular markers were used in this study to identify the potential molecular markers associated with anthracnose resistance traits in chili pepper cultivars. Among the four examined markers, HpmsE126 was detected in two anthracnose-resistant chili pepper cultivars, suggesting its close relation to the anthracnose resistance trait in chili pepper.
Conclusion: Given that two of the three most anthracnose-resistant cultivars, CN404 and TV3, possess HpmsE126 marker, this marker can be used to detect anthracnose-resistant lines in chili pepper breeding in Vietnam.
INTRODUCTION
In Vietnam, chili pepper is an important crop with high economic value. However, the annual chili yield is greatly affected by diseases caused by bacteria and fungi. One of the most well-known diseases in chili pepper is the anthracnose disease caused by the fungi species. These fungi were known to infect not only chili pepper but also other crops and orchard plants, such as apple, mango, coffee, etc. 1, 2, 3, 4. Multiple species have been found to be responsible for the anthracnose disease in chili pepper including , , , , , , and 5, 6. Apart from , , recent studies in Vietnam have revealed a number of species infecting chili pepper including, , , and 7, 8.
Among five domesticated chili pepper species, including , , , and , is the most commercially important species but is highly susceptible to anthracnose disease. The anthracnose resistance was introduced to by crossing with other Capsicum species 9, 10. The conventional breeding and selection processes of disease-resistant cultivars are often time-consuming and require financial resources for pathogenicity testing on large-scale. Different QTL mappings of anthracnose resistance were performed in the cross between and other Capsicum species discovered potential markers linked to anthracnose resistance trait 11, 12. The identification of anthracnose resistance-associated molecular markers allowed the marker-assisted or based breeding, which is less time-consuming and more cost-effective compared to conventional breeding 13, 14, 15. Given that anthracnose is one of the most destructive fungal diseases in chili-growing areas, including tropical Asian countries, e.g. Vietnam 16, thus the isolation of new resistant cultivars via molecular marker-based breeding will help to improve chili production and further bring economic benefits from chili pepper cultivation.
Therefore, the objectives of this study were (i) identifying the species majorly causing the anthracnose disease in chili pepper in Tra Vinh province (Vietnam), (ii) finding the cultivars showing resistance to these pathogens among commercial chili pepper cultivars in Vietnam, and most importantly, (iii) determining the potential anthracnose resistance-associated molecular markers which can be potentially applied further in chili pepper breeding in Vietnam.
MATERIALS AND METHODS
Plant growth condition
One wild bird-eye chili pepper line (TV3) found in Tra Vinh province and ten commercial cultivars (CN404, HNCS, CP01, HNN1, SGX5, SGX6, HN7777, TN535, HNNS, and PN400) were used in this study. Seeds from eleven cultivars were sown on small soil pots. At the 4-leaf stage, individual seedlings were transferred to pots with 18 cm in diameter. Plants were watered every morning with 2 l of water. Nutrients were supplied to each plant every two weeks with 250 ml of 1/10 MS (Murashige & Skoog) mineral solution. Plants were kept in a net house to avoid insects. Plants that were infected by mealybug were isolated and treated with insecticide (Bihopper 207EC, Binh Dien – Mekong limited company) until recovered.
Isolation of pathogenic fungi species
Fruits showing anthracnose symptoms (e.g., rounded necrosis spots on the fruit) were collected from chili pepper fields in Tra Vinh province. Infected fruit samples were washed five times by shaking in sterile water and dried under a laminar airflow chamber prior to surface sterilization with 70% ethanol for 2 min. The 0.5 cm x 0.5 cm fruit tissues containing the infected region were transferred to potato dextrose agar (PDA, 4% potato starch, 2% dextrose, 1.5% agar) plates kept at 25 C under 16 h light/8 h dark condition in an incubation room for seven days. Purified fungal strains were obtained after three consecutive sub-cultures on PDA plates. The pure fungal cultures were maintained on PDA plates for identification and pathogenicity test.
Morphological analysis of isolated pathogenic fungi species
Pure cultures isolated from infected fruits were first morphologically analyzed to identify the putative strains. The morphology of the fungi strains was analyzed by observing fungal colonies grown on PDA plates, conidia form, and the appressoria formation. For appressoria preparation, a small piece of thin PDA medium was prepared on a sterile glass microscopy slide. A small spore fragment taken from the fungal culture was distributed to four corners of the PDA medium prepared on the slide that was then sealed by a glass coverslip and placed on a sterile wet filter paper in a petri dish to maintain the humidity. After 14 days, the appressoria, which developed under the coverslips, were collected and placed on a glass slide for observation under a light microscope 1. The identification of putative strains was based on the description of Than . (2008) 17.
Identification of pathogenic fungi species via sequencing
Based on the morphological characterization, the putative samples grown on PDA medium were sent to the PHUSA Biochem LTD. company for sequencing, using the ITS1 primer (CTTGGTCATTTAGAGGAAGTAA). The sequenced ITS regions were used for the sequence homology analysis. To identify species, the sequencing results of the putative samples were used to BLAST with sequences available from GenBank Nucleotide database of NCBI (National Center for Biotechnology Information)
Pathogenicity test
The two isolated strains were used to study the anthracnose resistance of eleven chili pepper cultivars. isolates were incubated on PDA medium at 25 C for 14 days. To collect the fungal conidia, 5 ml of sterile water was added directly on the culture surface, followed by gently shaking of the petri dish. The conidia suspensions were collected to 1.5-ml tubes and centrifuged at 10000 rpm. The pellets that contained the concentrated conidia, was adjusted with sterile water to 10 conidia per ml using a hemocytometer.
For the pathogenicity test, the experiment was performed twice to confirm the phenotype. In each replicate, two to six fully developed green fruits from each cultivar were injected with 10 µl of the conidia suspensions (10 conidia per ml). Sterile water-infiltrated fruits were used as control. Three technical replicates were performed by injecting on three different places of a fruit of the cultivar that had medium or large fruits. To develop the disease, inoculated fruits were secured in plastic containers lined with wet paper towels and incubated for seven days at 25 °C. The development of disease symptoms was observed, and the lesion diameter was measured every 24 h. The lesion diameter was determined by averaging the largest and the smallest diameters of each lesion. The mean of lesion diameters at seven days after injection were used to determine the resistance of eleven chili pepper cultivars to the particular fungal isolate. Data of lesion diameter from two replicates were statistically analyzed using analysis of variance (P < 0.05), followed by multiple range tests using Tukey’s HSD test with SPSS 16.0 software (IBM).
Plant DNA extraction and PCR amplification
The genomic DNA of chili pepper was extracted using the CTAB method 18. The DNA quality and purity were determined by nanodrop (ND-2000, Thermo Scientific). To verify the presence of anthracnose resistance-associated markers in different chili pepper cultivars, polymerase chain reactions (PCRs) were performed using primers listed in
List of molecular markers and the respective primers used in this study
| Marker name | Primer sequence (5’ to 3’) | Product size | Reference |
| HpmsE032 | ATGCGCAAAGGGAGAAAATTCA | 282 bp | Suwor et al. (2017) |
| CGAACTAACCGTTCATGGTGGA | |||
| HpmsE116 | TCTCTCTCTACATCTCTCCGTTGA | 245 bp | Yi et al. (2006) |
| ACTCCCATACGGTGTCGTTC | Sun et al. (2015) | ||
| HpmsE126 | TGGGTATTTCCTTGCTGGAG | 108 bp | |
| TCCTTCCAGGAAACTGATGG | |||
| HpmsE047 | AACCCGTGTTCAATCCCCAAAT | 252 bp | Jayaram et al. (2015) |
| TGGCCATACCACCAGCAGTAGA |
RESULTS
Isolates obtained from infected fruits
From the infected fruits collected from chili pepper fields in Tra Vinh province, five different fungal isolates were obtained. Among these isolates, two putative strains, named 18T and 21T, were identified. The morphology analysis was performed according to the morphological descriptions of species outlined by Than . (2008) 17.
Colonies of the 18T strain grown on PDA showed pale orange color with fluffy aerial mycelia on the colony surface (Figure 1 A). Conidia of the 18T isolate were smooth-walled, transparent, colorless straight, cylindrical form with uneven ends. The size of the conidia was 10 – 14-µm long and 3 – 4-µm wide. The 18T appressoria were ovoid to ellipsoidal, 6 – 9-µm long, 4 – 6-µm wide, with medium to dark brown color. On the other hand, the colonies of the 21T strain grown on PDA were grey to pale orange with thin cottony mycelia on the surface (Figure 1 B). The 21T isolate produced fusiform conidia with both acute ends, 7 – 13-µm long and 3 – 4-µm wide. Most of the 21T appressoria were irregularly shaped, 5 – 8-µm long, 3 – 4-µm wide.
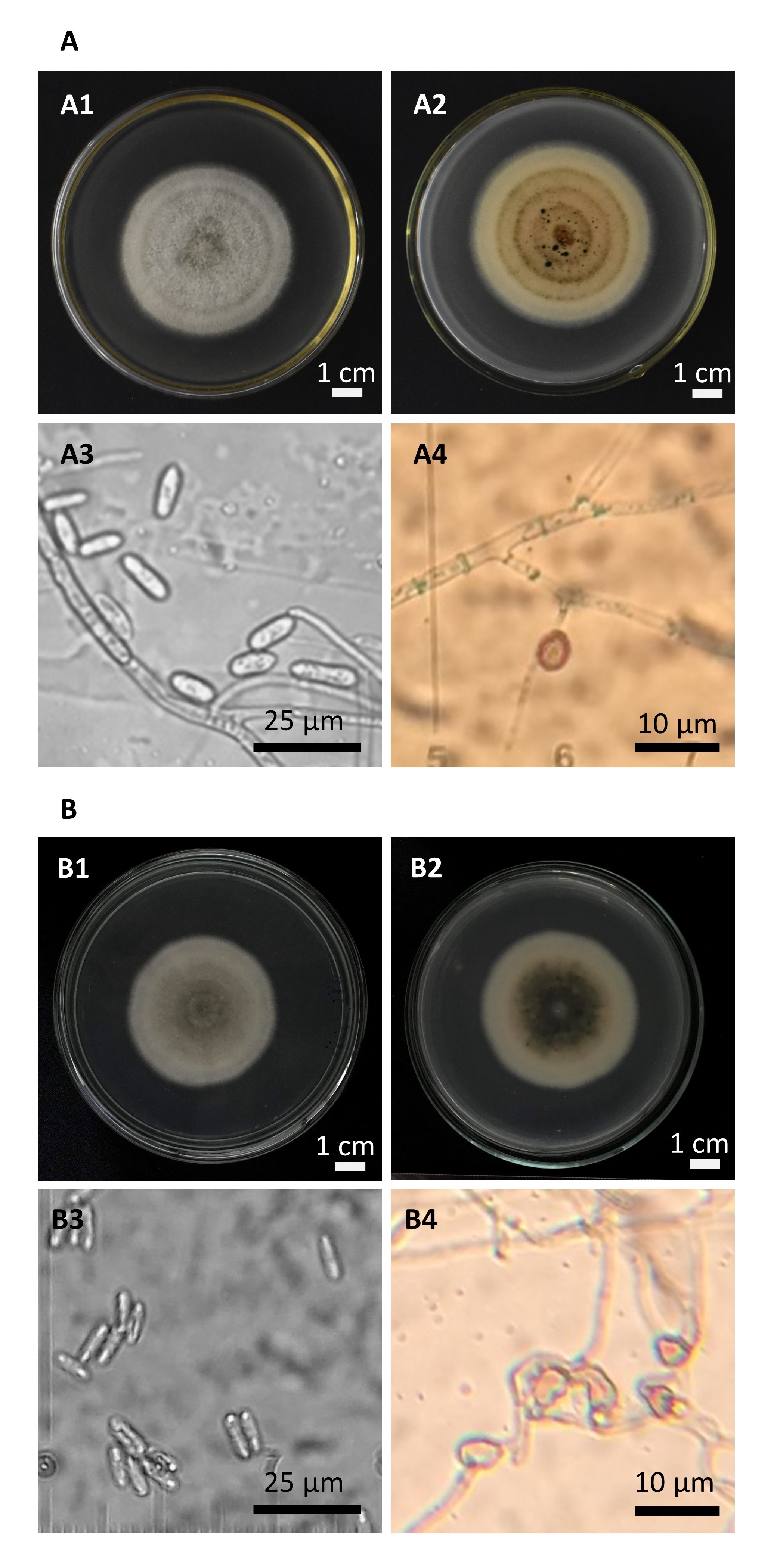
Morphology analysis of the
Morphology of the 18T isolate (A1 - A4) and the 21T isolate(B1 - B4). The morphological analysis based on the upper side (A1, B1) and lower side (A2, B2) of colonies grown on PDA medium, conidia (A3, B3) and appressoria (A4, B4).
The species of the two putative isolates were identified by sequencing the ITS region using the ITS1 primer. The sequencing result successfully confirmed the prediction that was based on the morphology observation. By comparing with the sequence database from NCBI, the ITS sequences of the 18T and 21T isolates showed the highest similarity to (Accession number: MH603141.1) and (Accession number: MF629920.1), respectively (data not shown). Therefore, together with the morphology observation, the 18T and 21T isolates were identified as and , respectively. These two fungal strains were later used in the experiment to determine the anthracnose resistance of different chili pepper cultivars.
Pathogenicity testing
Fruits from 11 chili pepper cultivars were infected with the two isolates of species, and . Both isolates were pathogenic and could successfully infect most of the cultivars. The infected areas were all rounded-shape with concentric acervuli (Figure 2 A). The lesion diameters of the eleven cultivars injected with the two isolates were described in table 2. The water-injected fruits significantly showed no lesion in eleven cultivars at seven days after injection (Figure 2). When injected with the conidia, the lesion diameters were all significantly larger than their corresponding water-injected controls (Figure 2 B). There were also significant differences in lesion diameter among eleven cultivars. The two cultivars, including CN404 and HNCS showed the strongest resistance to both species with the smallest infection diameter, smaller than 3 mm. In these cultivars, larger lesion diameters when infected by compared to that infected by indicated that they were more resistant to. Additionally, TN535 and HNNS were the two most susceptible cultivars to both species due to their large lesion diameters when infected by these two pathogens. The resistance level to was similar to in each cultivar, with the exception of the TV3 and PN400 cultivars (Figure 2 B). TV3 was strongly resistant to (3 mm in lesion diameter), but moderately resistance to (6 mm in lesion diameter) The PN400 cultivar, on the other hand, was susceptible to the (11.5 mm in lesion diameter) but showed the moderate resistance to (7.5 mm in lesion diameter).

Anthracnose symptoms on chili fruits from different cultivars at 7 days after injection. (A) Disease symptoms developed on chili fruits from eleven cultivars after injection with either
Potential anthracnose resistance-associated molecular markers
Four different PCR-based molecular markers (SSR markers), were tested to determine the potential molecular marker related to the anthracnose resistance in Vietnamese chili pepper cultivars. PCRs were performed for eleven chili pepper cultivars, and the summary of results was presented in table 2. Among the four tested molecular markers, HpmsE032, HpmsE047, and HpmsE116 were detected in both resistant and susceptible cultivars indicating these markers are not associated with the anthracnose resistance caused by defined species (Figure 3 B-D). Interestingly, the HpmsE126 marker was observed only in two of three anthracnose resistant cultivars, CN404 and TV3 (Figure 3 A). These two chili pepper cultivars carrying HpmsE126 showed very strong resistance to anthracnose since their lesion diameter was significantly lower than the lesion diameter in the cultivars without this marker. The data analysis also suggested that having HpmsE126 marker might contribute more to resistance than resistance (Figure 4).
Lesion diameter caused by two
| Cultivar | Lesion diameter* | HpmsE126 | HpmsE032 | HpmsE047 | HpmsE116 | |
| C. scovillei | C. acutatum | |||||
| CN404 | 2.04 a | 4.58 ab | X | X | X | X |
| HNCS | 2.67 ab | 3.25 a | X | X | X | |
| TV3 | 3.00 ab | 6.33 abc | X | X | X | X |
| CP01 | 5.86 abc | 6.28 abc | X | X | X | |
| HNNo1 | 7.28 abc | 9.13 cd | X | X | X | |
| SGX005 | 7.66 abc | 9.08 cd | X | X | X | |
| SGX006 | 8.02 bc | 8.44 bcd | X | X | X | |
| HN7777 | 8.47 c | 10.75 d | X | X | X | |
| TN535 | 9.42 c | 9.25 cd | X | X | X | |
| HNNS | 10.16 c | 10.75 d | X | X | X | |
| PN400 | 11.54 c | 7.50 bcd | X | X | X | |
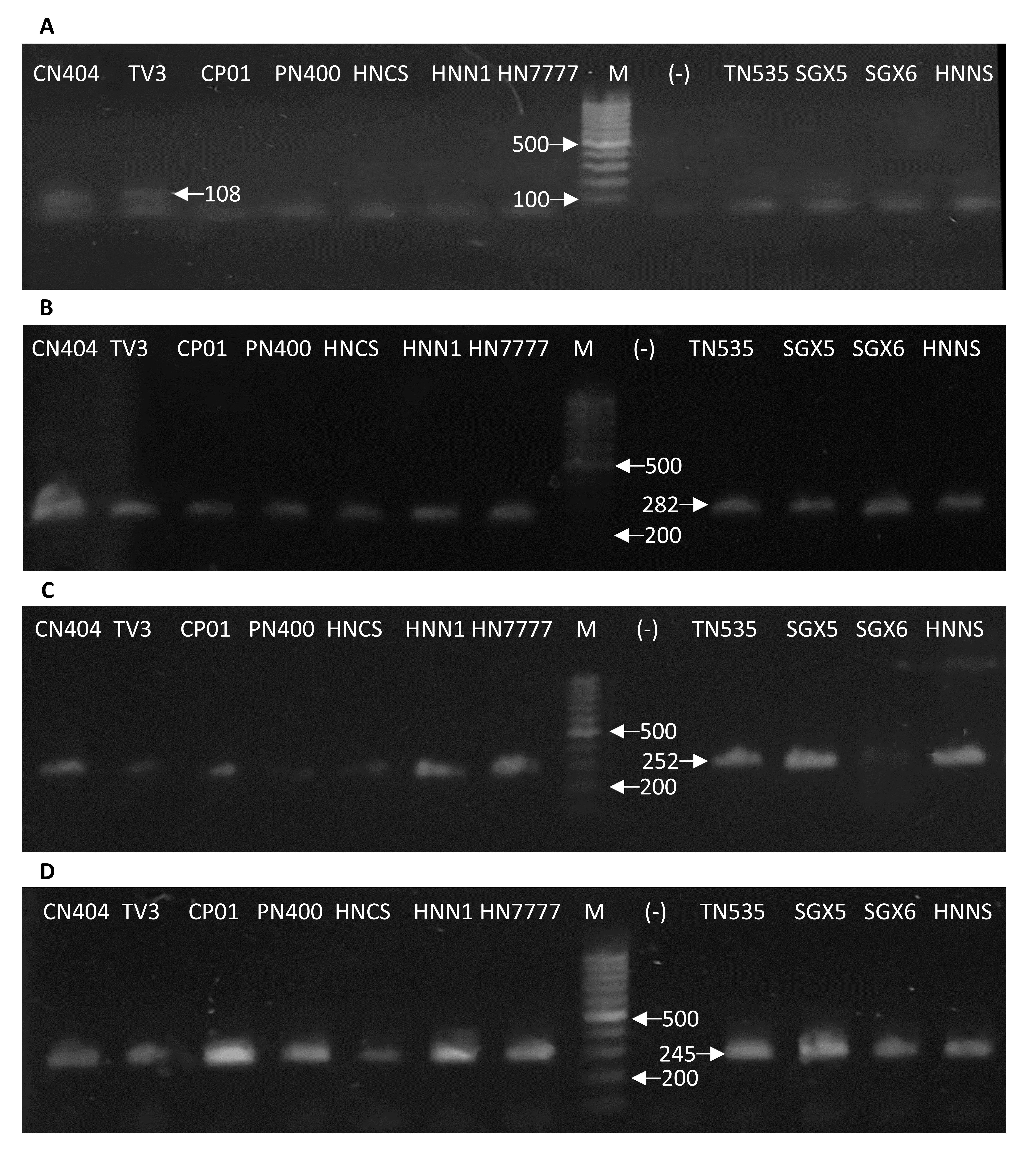
DNA electrophoresis of molecular markers amplified by PCRs in different chili pepper cultivars. Marker detection of HpmsE126 (A), HpmsE032 (B), HpmsE047 (C) and HpmsE116 (D) in eleven chili pepper cultivars. Lane M: 100-bp DNA Ladder, lane (-): DEPC-treated H2O. All PCR products were migrated in 2% agarose gel.
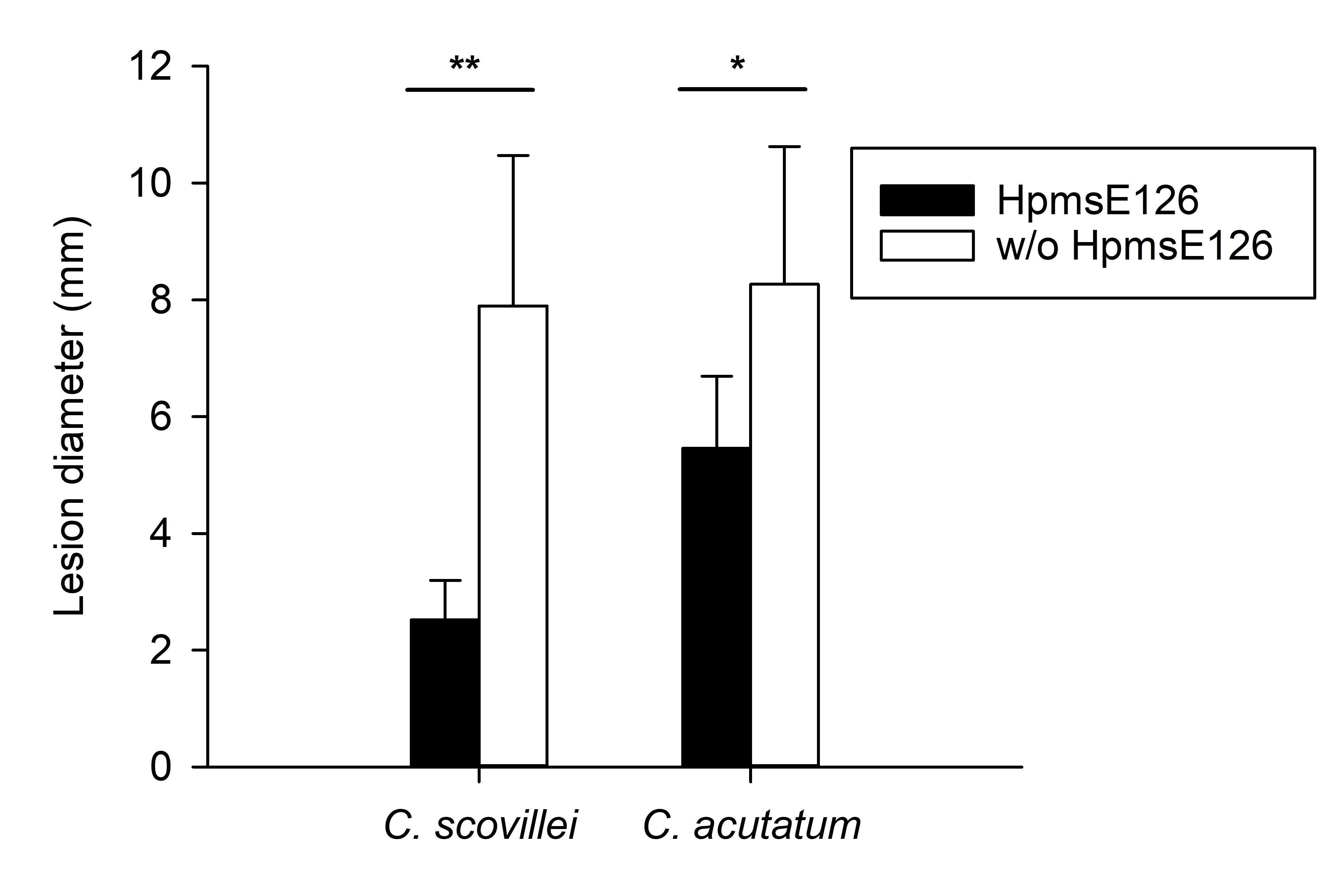
Comparison of lesion diameter between cultivars having HpmsE126 marker and cultivars without HpmsE126 marker. Data is presented as mean ± SD. Means were compared using Student’s t-test. Statistical difference is presented by either (*) (P < 0.05) or (**) (P < 0.01).
Among the markers showing differences between cultivars, HpmsE126 was observed in the two cultivars, CN404 and TV3, which had a very strong resistance to anthracnose, especially the disease caused by . As a result, the HpmsE126 might be related to the resistance to anthracnose and would be a potential molecular marker used in chili pepper breeding. Given that these chili pepper cultivars showed stronger resistance to compared to , the HpmsE126 marker might be more related to the resistance in chili pepper. Interestingly, it must be noticed that the HNCS cultivar was among the best anthracnose resistant cultivars in this study, although the HpmsE126 marker was not found in this cultivar (Figure 3 A).
DISCUSSION
In addition to isolation, which was previously identified in studies in Vietnam 7, 8, different species causing anthracnose disease in chili pepper in Vietnam, , was firstly identified in this study. This species was previously reported to cause chili anthracnose disease in Japan, Brazil, China, and Korea 6, 19, 20, 21. Recently, it was shown that in South Asia and South-East Asia, except Sri Lanka, was the prevalent anthracnose-causing pathogen and showed very high aggressiveness when infecting non-wounded fruits 22. In the study of Oo . (2017), all 36 tested cultivars belonging to , , and , were susceptible to the isolates 6. Crossing between and conferred to the resistance to in hybrid sweet pepper was reported in 2012 6. Interestingly, among the tested cultivars in this study, three cultivars, including CN404, HNCS, and TV3, showed strong resistance to the isolate obtained from Tra Vinh province. These cultivars could be a valuable resistance source for chili pepper breeding. According to the findings in this study, it is likely that the resistance trait has already been introduced into commercial chili pepper cultivars in Vietnam.
Most of the present commercial chili pepper cultivars belong to the species. While is commercially important, this species is highly anthracnose susceptible 23. From the 1990s, the potential anthracnose resistance sources were identified in other chili pepper species, including (PBC80, PBC81) and (PBC932) 9. In 2014, by studying the backcross BC population derived from the hybrid between line 77013 and the PBC932 using QTL analysis method, different markers, including HpmsE116 and HpmsE126, were identified to be closely related to the resistance of chili pepper fruit to 24, 25. In this current study, only the HpmsE126 was found to be related to moderate anthracnose resistance caused by . Interestingly, plants carrying this marker also showed additional strong resistance to , suggesting that a common mechanism might be shared between resistance and resistance. Although HpmsE126 was considered as a minor QTL related to anthracnose resistance, in the current study, this marker was more efficient in identifying the anthracnose-resistant cultivars compared to the other markers. Other markers, HpmsE032 and HpmsE047, were proven to be closely related to the anthracnose resistance 26, 27. However, in this study, these markers were not able to determine the anthracnose resistance variance in the tested chili pepper cultivars. It must be noted that the anthracnose disease in chili pepper is caused by different species. HpmsE047 was determined as a disease resistance marker when tested with and 26, 28. It is possible that chili pepper employs different molecular mechanisms to fight against different species. In the case of HpmsE032, although this marker was proven to be a -resistant marker, its efficiency was only 65% suggesting the involvement of other genes yet unidentified in the defence mechanism against and the combination of different marker would be more efficient in the detection of resistant cultivar 27. In 2011, Lee identified two markers related to the resistance to / (CaR12.2) and / (CcR9). The later marker was developed to SCAR marker (CcR9M1-SCAR) and was protected under the worldwide patent KR101010446B1 13, 28. These markers could be the potential additional tools for the detection of anthracnose-resistant cultivars, although its application in Vietnam could be difficult due to patent protection. In this current study, the strong resistance to both and of the HNCS cultivar without HpmsE126 marker supported the involvement of other gene clusters and other mechanisms in the resistance to and and the possibility to combine with other molecular markers in the determination of anthracnose resistance caused by and in chili pepper. The findings also support that the CN404 cultivar could be a very good commercial chili pepper cultivar for chili production and cultivation, especially in the regions affected by and . Additionally, the wild bird-eye chili pepper cultivar found in Tra Vinh province could be a potential anthracnose resistance source, which can be applied for chili pepper breeding.
CONCLUSION
In this study, from the infected fruits collected from chili pepper fields in Tra Vinh province, two isolates, and , were successfully isolated and identified using morphological and sequencing approaches. Among eleven tested chili pepper cultivars infected by these isolates, two cultivars were identified to have the resistance against both of these pathogens. Moreover, the marker analysis found the HpmsE126 SSR marker in the two resistant cultivars. The HpmsE126 might be an efficient molecular tool to identify the anthracnose-resistant cultivars for chili pepper breeding.
ABBREVIATIONS
ITS Internal transcribed spacer
SSR Simple Sequence Repeats
SCAR Sequence Characterized Amplified Region
NCBI National Center for Biotechnology Information
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
All the experimental works were performed by VA Ly. TPT Truong was involved in the pathogen isolation and molecular biology works. The project design and manuscript writing were performed by TH Nguyen.
ACKNOWLEDGEMENT
This research is funded by University of Science, VNU-HCM, under grant number T2018-17

