Immobilization of green fluorescence and lipase proteins in porous silica nanoparticles to improve their bioluminescence and catalytic activity
- School of Biotechnology, Department of Chemical Engineering, International University, Ho Chi Minh City, Vietnam
- Vietnam National University, Ho Chi Minh City, Vietnam
Abstract
Introduction: The GFP and lipase proteins have been immobilized in the aminated silica nanoparticles through encapsulation and ionic interaction. The synthesis of porous silica nanoparticles and amination of silica surface were simultaneously performed in a water-in-oil emulsion.
Methods: The materials have been characterized by zeta potential measurements, electron transmission microscopy, and fluorescence spectroscopy. The reaction mixture was analyzed by gas chromatography equipped with a mass selective detector.
Results: The aminated-silica nanoparticles with a positive zeta potential interact with GFP having a negative zeta potential via ionic force. GFP immobilized by the aminated silica nanoparticles greatly enhances (ca. 2 times) the bioluminescence intensity compared to free GFP. The lipase/aminated-silica nanoparticles catalyzed the transesterification reaction of 1-hexanol and ethyl acetate. This catalyst shows better activity than free lipase and remains catalytic activity over at least eight catalytic cycles in an organic solvent.
Conclusion: The result shows that the immobilization of proteins in the aminated-silica nanoparticles improves their bioluminescence and catalytic activity.
INTRODUCTION
The fluorescent proteins (FPs) were discovered in the early 1960s by Osamu Shimomura during his study on identifying the glow of jellyfish from . FPs have a wide range of applications such as reporter assays 2, 3, fluorescence microscopy 4, 5, 6, analysis of the colocalization of proteins7, macro-photography 8, and transgenic pets 9. FPs have been immobilized into solid materials to fabricate biosensors and optoelectronic surfaces. There are some applications of FPs attached to solid materials such as pH sensors 10, sensors for monitoring mechanical failure11, toxicity biosensors 12, 13, pH sensors for biocatalytic study 14.
Lipases play an important role in industrial biocatalysis that takes place in both aqueous and non-aqueous reaction media. Moreover, they are well known for many organic transformations such as chemo-, region-, and enantio-selectivity. Many studies tried to modify the structure of lipases to make them suitable for reactions in the organic solvent. However, those modifications are expensive, and hence it is better to find the right lipases that are tolerant with organic solvent 15, 16. However, in reality, most lipase enzymes are denatured and become inactive in the organic solvent. Therefore, protein engineering, chemical, and physical methods, such as immobilization of lipases proteins in solid materials, are needed to improve their stability in the organic solvent 17, 18, 19.
Immobilization of proteins in inert and insoluble supports is of great interest concerning scientific and industrial aspects. Compared with the soluble form of proteins, the immobilized proteins possess many improved properties, including i) being easy for workup because of a negligible amount of proteins in the reaction mixture; ii) being simple for recovery via centrifugation or filtration and reuse them; and iii) improving the thermal stability and used conditions 20. There are typically two approaches to immobilize proteins in supports: irreversible and reversible immobilization. However, these two approaches having encountering objectives - stability and reversibility - are difficult to fulfill simultaneously.
The irreversible immobilization of proteins includes forming covalent bonds, encapsulation or micro-encapsulation and cross-linking. On the contrary, the reversible immobilization of proteins is performed via ionic binding, adsorption, chelation, affinity binding, and disulfide bonds. The biggest disadvantage of the latter approach is the leakage of proteins from the supports; however, the decayed proteins can be reloaded with fresh proteins 21. In general, the immobilization of proteins results in loss of catalytic activity and mass transfer limitation; however, the half-life is significantly improved20, 22, 23.
Regarding the catalytic application of proteins, it is desirable to make proteins recycle and stable. To this end, irreversible immobilization is the most interesting approach. If proteins firmly interact with supports via reversible immobilization, they are readily detached from the supports during chemical reactions in the liquid phase. Furthermore, the change in reaction conditions such as pH, temperature, and ionic strength also promote the detachment of proteins from supports 23, 24. Ideally, once proteins are attached to supports using irreversible immobilization, they cannot be detached without destroying either the biological activity of proteins or the supports.
This study immobilized GFP and lipase proteins in porous silica nanoparticles by encapsulation and ionic interaction. Both encapsulation and ionic interaction between proteins and silica surfaces have been achieved by synthesizing porous silica nanoparticles and aminated modification of silica surfaces during the immobilization process in a water-in-oil emulsion. The GFP and lipase were employed in this study to show the potential application of our method for several proteins.
MATERIALS - METHODS
Chemicals
Triton X-100 (Merck, 95.0%), cyclohexane (Merck, 99.5%), hexanol (Merck, 99.0%), ammonium hydroxide (Merck, 28.0%), green fluorescence protein (Merck, Mw ~ 28 kDa), tetraethyl orthosilicate (Alfa Aesar, 99.0%), (3-aminopropyl)triethoxysilane (99.9%, Merck), ethanol (95.0%, Alfa Aesar), lipase from Candida Antarctica (lyophilized, powder, beige, ~0.3 U/mg, Merck).
Immobilization of GFP in aminated-silica using reverse micelle approach
The water-in-oil emulsion was created as follows: mixing Triton X-100 (10 g) with cyclohexane (30 mL) and hexanol (8 mL) to have the solution A. Hexanol was added as a co-surfactant to produce reverse micelles. Water (2 mL) with different pHs (pH = 9.2, 10.2, 10.9, 11.9, or 12.4 was adjusted by NHOH solution and measured by a pH meter) was added to solution A to have a water-in-oil emulsion. The emulsion was stirred at 550 rpm for 5 min at 25 C, and then GFP in water (dialyzed, 50 L, 6 mg/mL) was added. TEOS (100 L) was added to the above emulsion containing protein in the water pool to perform the immobilization of protein in silica. The hydrolysis and condensation of TEOS were lasted for 16 h at room temperature at a stirring rate of 550 rpm. At this reaction time, vials with water pools having pH = 9.2, 10.2, and 10.98 do not appear precipitate of silica. Therefore, an aqueous solution of NaF 1M (50 L) was added to those vials to speed up the rate of TEOS hydrolysis and condensation reactions. After an additional 2 h reaction, 200 L of APTES (200 L of pure APTES in 1.4 mL of ethanol) was added to all vials, and the reaction was lasted for an additional 25 h. Ten milliliters of ethanol were added to each vial to destabilize emulsion and to precipitate nanoparticles. The colloidal solution was centrifuged at 5000 rpm for 30 min to retrieve the nanoparticles. At this stage, the color of nanoparticles was switched from green to white after centrifuging. The residue was redispersed in 30 mL of water and then centrifuged at 500 rpm for 30 min. The green color of nanoparticles reappeared. The nanoparticles were washed twice with 40 mL of water. The final nanoparticles were dispersed in 40 mL of water. The GFP/aminated-silica nanoparticles were freeze-dried using a lyophilizer.
Immobilization of lipase in aminated-silica using reverse micelle approach
One hundred grams of Triton X-100 was added to 300 mL of cyclohexane and 80 mL of hexanol to have the solution A. Fifteen milliliters of water were added to solution A to have a water-in-oil emulsion. This emulsion was stirred at 550 rpm for 1 h before 1 mL of aqueous lipase (10 mg/mL) and 1 mL of TEOS were added. The solution was stirred for an additional 1 h before 5 mL of NHOH (28%) was added to hydrolyze TEOS. The hydrolysis and condensation reactions were lasted for 20 h, and then 1 mL of APTES (400 L of APTES in 2.8 mL ethanol) was added. The reaction was stirred for additional 6 h. The lipase/aminated silica nanoparticles were purified by washing with ethanol and water several times and then redispersing them in water or freeze-dried them using a lyophilizer.
Characterizations
The measurements of fluorescence spectra were performed with a fluorescence analyzer (HORIBA); the exciting wavelength for GFP was set at 395 nm. The zeta potentials of the samples dispersed in deionized water were measured using a Malvern Zetasizer Nano ZS instrument. The folded capillary cell (DTS1060) was first flushed with ethanol to wet the inner surface of the cell. It was then flushed with water to replace the ethanol residue before the colloidal samples were introduced into the cell using a syringe. The reported zeta potentials are at 25 C and neutral pHs. Samples were characterized by transmission electron microscopy (Titan G2 80-300 kV, FEI company, 4 x 4 x 4 k CCD camera).
The catalytic activity of lipase immobilized in silica was tested for the transesterification reaction between ethyl acetate and 1-hexanol. In a typical reaction, 10 mg of lipase immobilized sample was dispersed in 10 mL of acetone containing 100 mM ethyl acetate and 100 mM hexanol. The mixture was stirred at 30 C for 24 h. The conversion and product distribution of the reactions catalyzed by enzyme catalysts were determined by gas chromatography coupled with a mass analyzer (Agilent 7820A-5975C). The non-polar column DB-5MS (30 m x 0.25 mm x 25 mm) was used to separate the reaction products.
RESULTS
Zeta potential measurements were measured for GFP, aminated-silica, GFP/aminated-silica (pH 9.2), GFP/aminated-silica (pH 10.2), GFP/aminated-silica (pH 11.9), and GFP/aminated-silica (pH 12.4) samples. The resulting zeta potentials of those samples are shown in Figure 1 and
The digital images of the GFP/aminated-silica samples dispersed in water and dried with a lyophilizer are shown in Figures 2a and b. The samples in Figure 2 were exposed under a UV light at a wavelength of 365 nm. The samples synthesized using the water pools at pH9.2 and pH11.9 show a brighter green color of GFP than those at pH10.2 and pH12.4. This observation relates to the stability of GFP under different pH values during the synthesis of the GFP/aminated-silica samples and also to the structure of the aminated-silica samples. Our observation agrees with the previous study showing that free GFPs are not stable under an acidic environment and are highly stable at alkaline pH 26.
The numerical values of zeta potentials of the samples
|
Sample |
Aminated-silica |
GFP |
GFP/aminated-silica (pH9.2) |
GFP/aminated-silica (pH10.2) |
GFP/aminated-silica (pH11.9) |
GFP/aminated-silica (pH12.4) |
|
Zeta, mV |
+31.4 (0.7)* |
-23.5 (9.6) |
-28.8 (1.2) |
-16.5 (2.9) |
-22.1 (1.1) |
-26.5 (5.0) |

The measurements of zeta potentials of aminated silica (back), GFP (olive), GFP/aminated-silica synthesized at pH9.2 (pink), GFP/aminated-silica synthesized at pH10.2 (red), GFP/aminated-silica synthesized at pH11.9 (blue), and GFP/aminated-silica synthesized at pH12.4 (magenta).
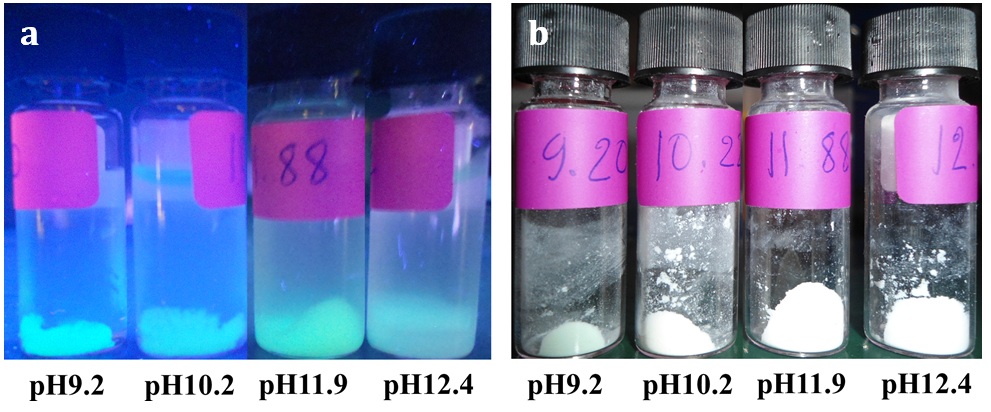
The digital images of the GFP/aminated-silica samples dispersed in water (a) and being dried with a lyophilizer (b) exposed under a UV light with a wavelength of 365 nm.
The measurements of fluorescence spectra of the samples were performed using a fluorescence analyzer with an exciting wavelength for GFP at a wavelength at 395 nm. The fluorescence spectra of the GFP/aminated-silica samples dispersed in water (after purification) and the freeze-dried samples (after purification) dispersed in water are shown in Figure 3.

The fluorescence spectra of the GFP/aminated-silica samples dispersed in water at pH7.0 (a) and after freeze-dried and redispersed in water at pH7.0 (b).
For the GFP/aminated-silica samples dispersed in water, the emission intensity of the samples synthesized using the water pools at different pH values is ranked as follows: pH11.9 > pH9.2 > pH10.2 > pH12.4 > free GFP. This observation is consistent with what we observe from the digital images showing in Figure 2. For the GFP/aminated-silica samples after being freeze-dried and redispersed in water, the emission intensity of those samples declines as shown in Figure 3b. The GFP in the sample synthesized with the water pool at pH9.2 becomes brightest because its emission intensity is highest among the samples.
To reveal the structure of aminated-silica and GFP/aminated-silica nanoparticles, the TEM measurements were performed. The TEM images of those samples are shown in Figure 4. The structure of the control sample (aminated-silica) is shown in Figures 4a and b with different magnifications. The porosity of the aminated silica sample is also not visualized from the TEM images. To ensure the nanoparticles in Figure 4b captured by TEM are nanoparticles consisting of silicon element, the silicon elemental analysis was performed. The silicon elemental analysis is shown in Figure 4c. The white areas in Figure 4c indicate the presence of silicon elements in nanoparticles.
Figures 4d, e, and f show the structure of the GFP/aminated-silica nanoparticles synthesized at pH10.2, pH11.9, and pH12.4, respectively. The size distribution of those nanoparticles is also not uniform because of the nature of water droplets in the oil phase. However, the porosity of those samples, indicated by white spots inside each nanoparticle, is better than that of the aminated silica sample. The better porosity of those samples than the aminated silica sample is attributed to the templating role of GFP. The porosity of those samples was created and observed during the TEM characterization. This is due to the soft nature of GFP, which is sensitive and not stable under the electron beam.
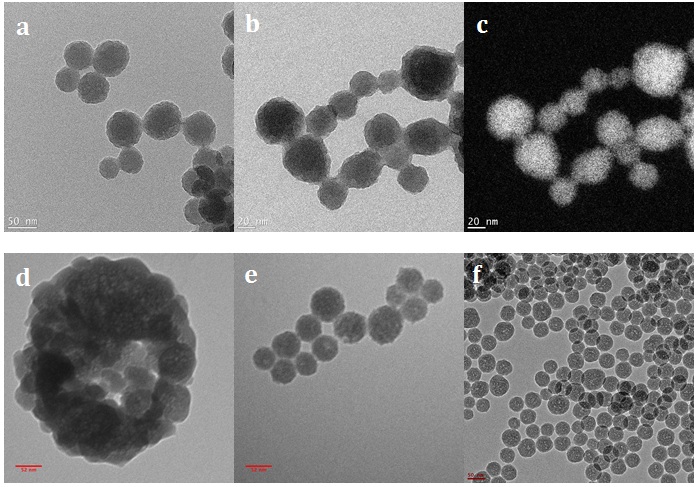
The TEM images of the aminated-silica samples were captured at different magnifications (a, b) and the silicon element mapping (c) from image (b). The TEM images of the GFP/aminated-silica samples were synthesized at pH10.2 (d), at pH11.9 (e), and pH12.4 (f). Those samples were dispersed in the water right after synthesis and purification. The scale bars for (a), (b), (c), (d), (e), and (f) sub-images are 50, 20, 20, 52, 52, and 50 nm, respectively.
The lipase/aminated silica nanoparticles were synthesized following the procedure described in the experimental section. The free lipase catalyst was tested for the transesterification reaction between 1-hexanol and ethyl acetate, but GC/MS did not detect the hexyl acetate product. This observation indicates that the free lipase was not stable in a mixture of the organic reaction medium. On the contrary, the lipase/aminated-silica sample shows the product in the reaction mixture (Figure 5b). The catalytic activity of the lipase/aminated-silica sample is decreased after the first catalytic cycle (Figure 5a). However, its activity remains stable from the second to eighth catalytic cycle.
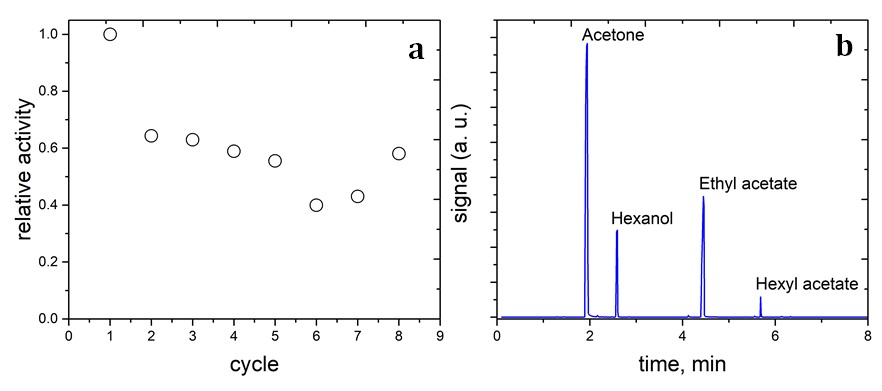
The relatively catalytic activity of the lipase/aminated-silica sample for the transesterification reaction between 1-hexanol and ethyl acetate (a) and the chromatogram of the reaction mixture (b).
DISCUSSION
The purpose of the surface modification of silica nanoparticles with APTES is to convert its negative zeta potential to positive zeta potential. This, along with the physical immobilization of GFP by porous silica, ensures to enhance the ionic force between GFP with negative zeta potential (-23.5 mV) and aminated-silica surface with positive zeta potential. As seen in
The suitable pH for the immobilization of GFP in the aminated silica nanoparticles is around pH11.9 because the fluorescence intensity of GFP in this sample is higher than that in other samples. Moreover, all immobilized GFP/aminated silica samples show better fluorescence intensity than free GFP. Therefore, the GFP/aminated-silica nanoparticles are potential fluorescence probes because GFP and its variants are widely used in cell and molecular biology for in vivo imaging and probing. The observation relates to the denaturation of GFP during the freeze-drying process, and this denaturation is not reversible once GFP was redispersed in water. Our observation suggests that the GFP/aminated silica nanoparticles should be stored in the water right after synthesis and purification to preserve their bioluminescence.
The particle size distribution of the aminated-silica and the GFP/aminated-silica samples observed by TEM is not uniform because it was synthesized using the water-in-oil method. In this method, water was dispersed in cyclohexane solvent by mechanical stirring, and the water droplets were stabilized by Triton X-100 surfactant. After that, the silica precursor (TEOS) was added and was under hydrolysis and condensation in water droplets. Therefore, the size of nanoparticles was determined by that of water droplets. However, the characteristic feature of the size distribution of water droplets is not uniform in the oil phase 27. As a result, the size distribution of aminated silica nanoparticles is not uniform.
The loss of catalytic activity at the second cycle is referred to the loss of catalyst during the recovery process and/or to the leaching of the weakly-bound-lipase with the aminated-silica surface. In summary, in comparison with the free lipase, the immobilization of lipase with aminated silica nanoparticles greatly improves the stability of lipase and also remains its catalytic activity. The structural complexity of lipase enzyme and/or the diffusibility of organic reactants could be altered after several catalytic cycles. As a result, this change in the structure of the enzyme and/or diffusibility of molecules increases the catalytic activity from the sixth to the eighth cycle.
CONCLUSIONS
We have successfully immobilized GFP and lipase in the aminated silica nanoparticles. Compared with the free GFP, the GFP/aminated silica nanoparticles significantly enhance the bioluminescence of GFP. The lyophilization (freeze-drying) to remove water from the GFP/aminated silica samples are not recommended because this process results in the loss of the bioluminescence of GFP. The best pH value of the water pool for the synthesis of GFP/aminated silica nanoparticles is 11.9. The immobilization of lipase in the aminated silica nanoparticles enhances the catalytic activity for the transesterification reaction between 1-hexanol and ethyl acetate in an organic solvent and keeps the catalytic activity of the lipase protein stable over at least eight catalytic cycles.
LIST OF ABBREVIATIONS
GFP: Green fluorescence protein
TEOS: Tetraethyl orthosilicate
APTES: (3-Aminopropyl)triethoxysilane
GC/MS: Gas chromatography/mass spectrum
TEM: Transmission electron microscopy
COMPETING INTERESTS
The author(s) declares that they have no competing interests.
ACKNOWLEDGMENT
This research is funded by International University, VNU-HCM under grant number T2020-04-BT.
AUTHORS CONTRIBUTIONS
Khanh Bao Vu: Conceptualization, Investigation, Methodology, Funding acquisition, Writing manuscript. Thanh Khoa Phung: Conceptualization, Methodology, Experiment. Thong Hoang Le, Khoa Dang Tong, Huy Hoang: Experiment. Thao Huynh Thanh Nguyen, Linh Thi Bui: Experiment and Editing. All authors read and approved the final manuscript.

