Rapid sex determination of preimplantation bovine embryo using direct-PCR from a single blastomere
- School of Biotechnology, International University - Vietnam National University HCMC, Ho Chi Minh 700000, Vietnam
- School of Biotechnology, International University - Vietnam National University HCMC, Ho Chi Minh 700000, Vietn
Abstract
Introduction: Sex control plays an indispensable role in livestock farm management. It enables farmers to quickly respond to the needs of herd rearrangements such as size and composition, and stably maximize the economic value, particularly on long gestation period animals as bovine. Despite being an invasive method, genetic examination of blastomeres isolated from the preimplantation embryos provides a superior accuracy method for bovine sexing, particularly with a highly sensitive molecular technique such as PCR. Currently, this technique is commercially expanding due to many improvements in embryo technology. In this study, we attempted to reduce the embryonic invasiveness and develop a simple, fast, and reliable sexing procedure using direct amplification of Y chromosome-specific marker from only one blastomere.
Methods: The bovine sexing was carried out using direct PCR of Y chromosome amplicon from single blastomeres biopsied from sex-known somatic nuclear transfer technology (SCNT) embryos (male) and enucleated metaphase II nucleus (female). The intensity of the amplified products was also optimized using the PCR additive.
Results: Utilizing the protocol resulted in the success of sex typing for all tested samples in this study that are comparable with genomic DNA amplification.
Conclusion: Our result could serve as a fast procedure for bovine embryo sexing and would be an effective tool for sex ratio control in the husbandry industry, animal embryo research, and endangered animal preservation.
Introduction
It is crucial and efficient to manage the animal herd in the livestock industry through sex planning1. This management is benefit not only because of the economic value difference between sexes but also the capability that enables the livestock producer to fast respond to increasing the size of the herd, especially in the case of long gestation period species as bovine2.
There are several approaches for bovine sex control, and each one has its advantage and shortcoming. The most common, commercially successful method is using flow cytometry sex-sorted semen for artificially inseminate into female bovine 3, 4. Although the accuracy could reach ninety percent of the pregnant cases, this technique persisted several drawbacks such as higher cost of sorted semen, reduced semen performance, and lower embryo yield fertilization (IVF), compared with unsorted semen5. Other methods such as ultrasonography and Y chromosome fetal DNA detection from pregnant female plasma were non-invasive and accurate6, 7. However, the techniques could be conducted only when the pregnancy was already established and having no preselection value.
Recently, many improvements in embryo technologies have enabled the commercial expansion of embryo-based sex determination, including embryo manipulation, preimplantation genetic diagnosis, cryopreservation, and embryo transfer8, 9, 10, 11, 12. Using a genetic examination method from limited embryonic biopsied blastomeres allows the technician to select the highest quality embryos and sex determination before transfer to the synchronous recipient cows. The embryo techniques are also considered a meaningful tool for preserving endangered animal species. The selecting possibility of the sex of the offspring may have a significant impact in the breeding program where each male can mate with several females to speed up the propagation process13, 14. Among the molecular methods applied on the collected blastomeres, cytogenetic techniques such as karyotyping or fluorescence in situ hybridization (FISH) are laborious, time-consuming, and expensive compared with PCR which was also highly accurate and robust15.
Nonetheless, to improve the diagnosis reliability using PCR, more blastomeres also need to be collected to increase the sensitivity, leading to the concern about the quality and viability of the biopsied embryos16. Therefore, this study attempted to develop a simple, fast, and highly sensitive method for bovine embryos sex determination using direct PCR with a single embryonic blastomere. In addition, we also discussed the potential of the method for enrichment of genetic typing for embryo selection, which is advantageous for the husbandry industry and endangered animal preservation programs.
Materials and methods
Genomic DNA extraction
Bovine genomic DNA was extracted from marketed beef using the standard protocol17. Briefly, 0.5 g of bovine meat was collected and incubated at 55°C for six hours in the lysis buffer (10 mM Tris–HCl pH 8.0 and 0.1 M EDTA) containing 0.5% SDS and 20 μl of 20 mg/ml proteinase K (Promega, Madison, WI). Next, the supernatant was purified with phenol/chloroform/isoamyl alcohol extraction, and DNA pellets were obtained by alcohol precipitation. After being dissolved in water, the DNA was quantified and qualified by NanoDrop One spectrophotometer and agarose gel electrophoresis (ethidium bromide-stained and visualized under UV light using Gel-Doc-It 310 Imaging System – Upland, CA, USA).Then, the DNA concentration was adjusted to 50 ng/μl, which is suitable for PCR.
Preparation of single-cell and single blastomere
Primary fibroblast cells were isolated and cultured from a male cow lung tissue collected from a local slaughterhouse using a standard protocol18. Then, the cells were cultured in DMEM high glucose media (Hyclone, UT, USA) supplemented with 10% fetal bovine serum (FBS, Hyclone), 1% penicillin-streptomycin (Gibco, NY, USA), and 2 mM L-glutamine (Gibco) at 38.5 °C, 5% CO in a humidified incubator (Panasonic, Japan). For single-cell collection, 80 – 90 % confluent cultured disk was trypsinized, FBS neutralized and harvested in a 15 ml centrifuge tube. Next, the cell suspension was washed twice with Phosphate-buffered saline (PBS). Then, the single cells were observed under a stereoscopic microscope and transferred into a 0.2 ml PCR tube using a 30-µm diameter glass mouth pipette.
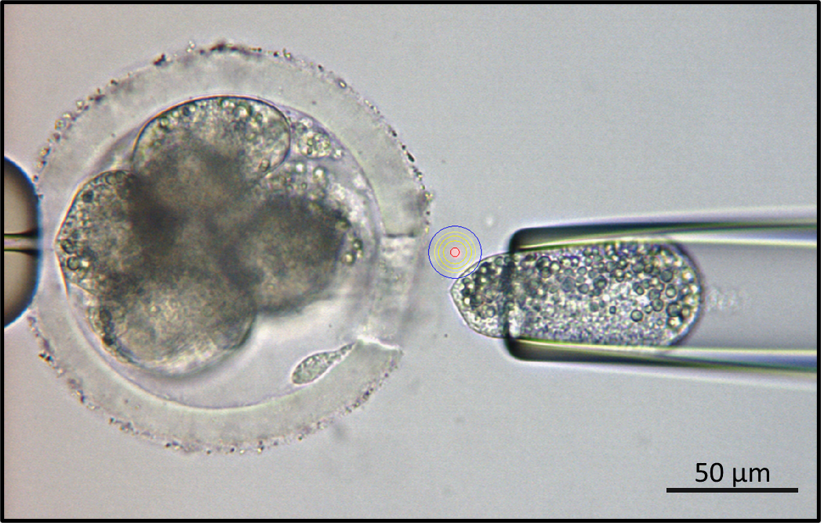
Bovine 8-cell embryo biopsy with the assist of XYClone laser system. Bar scale indicated for the microscopic magnification.
For single blastomere collection, bovine somatic cell nuclear transfer (SCNT) embryos were kindly prepared and provided by Cell Reprogramming Laboratory (International University, VNU) using established protocol 19. Briefly, oocyte-cumulus-granulosa complexes (OCGs) were isolated by aspiration from large antral follicles (4-7 mm in diameter) using an 18-gauge needle. The OCGs were cultured to the first polar body extrusion to the maturation stage and reached the metaphase II phase. After that, the metaphase II chromosome of matured oocytes was taken out, and the bovine fibroblast donor cell prepared as described previously was injected into enucleated oocytes using a micromanipulator system (Nikon Eclipse Ti-0, Japan). The reconstructed oocytes were artificially activated by Calcium Ionomycin for 5 minutes following by 4 hours of 6-dimethynopyridine (6-DMAP) in the modified synthetic oviduct fluid (mSOF) medium to produce diploid embryos. Then the cloned embryos were cultured in mSOF medium for seven days to the blastocyst stage. Twenty blastomeres were collected from the 8-cell (2 embryos) or 16-cell embryos (1 embryo) stage using XYClone laser system (Hamilton Thorne — USA), which integrated with micromanipulator system (Figure 1), and then were kept into 0.2 ml PCR tubes. For female single-cell sexing, five metaphase II chromosomes (2n-nucleuses) enucleated from the matured oocytes were also collected into the 0.2 ml PCR tubes for the direct PCR reaction. All the samples were stored at -20C until use.
Direct PCR
Amplification for bovine sexing was indicated by an amplicon specific for Y chromosome (BY) with the size of 300 bp using primer pair BY-F 5’-CTCAGCAAAGCACACCAGAC-3’ and BY-R 5’-GATCTGTAACTGCAAACCTGGC-3’ 20, 21. For PCR-sexing, amplification reactions were performed in 20 μl containing 50 ng genomic DNA, 0.5 μM of each primer, 200 μM dNTPs, 1X PCR buffer, and 0.5 U of FU14 DNA polymerase (Salagene, HCMC, Vietnam) using a PTC-100 Thermocycler (Bio-rad, USA). Cycling profiles for PCR consisted of an initial denaturation at 95C for 5 minutes followed by 35 cycles of 20 seconds at 95C, 50 seconds at 52C, 20 seconds at 72C, and a 5 minutes final extension at 72C. Ten μl of PCR products were checked by electrophoresis on 2.5 % agarose gel in 1 X Tris-acetate-EDTA (TAE) buffer. After ethidium bromide staining, the gel was visualized under ultraviolet (UV) light.
Direct PCR for single-cell and single blastomere
For direct PCR-sexing on the single cell, a master mix containing BY primers was added into a 0.2 ml PCR tube containing a single somatic cell or embryonic blastomere at the final concentration and cycling condition as same as in genomic DNA amplification. For improving the assay sensitivity, ammonium sulfate was added to the reaction at a final concentration of 0, 1.25, 2.5, 3.75, and 5 mM to find the optimal one. Ten μl of PCR products were visualized by 2.5 % agarose gel electrophoresis under UV light for sex determination. For detail, 300 bp amplicon indicates the male embryo and vice versa; the absence denotes the female one.
The method accuracy was accessed via the sensitivity and specificity showed by the percentage of successful diagnosis of the male from sex-known SCNT blastomeres (male) and female from enucleated metaphase II nucleus (female), respectively.
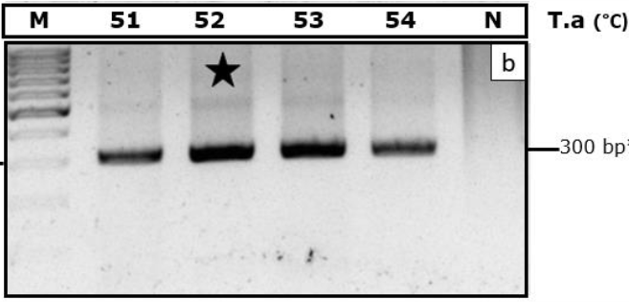
Optimization of PCR using male genomic DNA for and BY) marker. T.a indicates annealing temperatures (oC). M, DNA ladder100 bp. N indicates negative control. The dark star indicates optimal temperature.
Results
Optimization of PCR for bovine sexing
For sex determination in bovine at the single-cell level, we first selected several candidate amplification markers that were successfully established previously 1. Then, we chose some potential candidates based on the practical success of bovine sex determination in various reports. As showed in Figure 2, male genomic DNA amplifications were successful on BY marker with 300 bp product, without any considerable non-specific amplified products. The PCR result also showed that primer pairs specifically amplified from 51C to 54C for the annealing step, 52C was optimal, showed by electrophoresis band intensity. After that, this PCR condition was set for the next experiment.

Optimizing of bovine sex determination using direct PCR from single blastomere with PCR additive. M, DNA ladder. AS, ammoniumsulfate used at different concentrations. gDNA, amplified from genomic DNA. 1,denoted for single blastomere amplification, ♂ male cell, ♀ female cell. N, negative control. The dark star stands for optimal concentration of ammonium sulfate.

Sex determination of bovine embryo using direct PCR from single blastomere. M, DNA ladder. gDNA, amplified from genomic DNA. ♂, Malecell. ♀, Enucleated metaphase II from the bovine oocyte. N, negative control.
Enhance sensitivity of sex determination of bovine embryo using direct PCR from single embryonic blastomere.
The most crucial challenge of single-cell amplification is the limited amount of genetic material as the template for PCR compared with amplification using extracted genomic DNA, even down to a single copy as a Y chromosome from one male cell. For that reason, we excluded the DNA extraction step out of the sexing process, which introduces the risk of loss of DNA material. Because of embryonic blastomere quantity limitation, we first optimized the protocol using single somatic cells, which were assumed to possess similar contents. As expected, although it was successful in amplification, however, the band intensity was much lower compared with genomic DNA when using the single blastomere (Figure 3) or single somatic cell (data not showed).
To enhance the method sensitivity, we utilized ammonium sulfate as the PCR enhancer, well-known for destabilizing effect, preventing mismatched primer-template base-pairing, thereby enhancing specificity and PCR yield22, 23. As showed in Figure 3, the amplification of BY from the single blastomere was improved when increasing ammonium sulfate concentration. The result from a single blastomere at an optimal concentration of 5 mM was comparable with amplification from genomic DNA. However, a higher ammonium sulfate concentration did not show higher PCR efficiency (data not shown). In female blastomere, there was a faint non-specific amplicon, which did not affect the sex conclusion. Therefore, the method successfully discriminated between males and females at the single-cell level for all tested samples (Figure 4) with 100 % sensitivity and specificity.
Discussions
Detection of the presence of the Y chromosome from embryonic blastomere using a sensitive molecular technique as PCR provided a reliable, accurate method for bovine embryo sexing, compared to the other ones. However, in many cases, several blastomeres from one embryo were collected for the diagnostic process to ensure the sensitivity and prevent the false negative recognition of male genetic factors16. Additionally, it was reported that the difference between the biopsied group and the control group in the embryo viability was not significant24. However, there were also reports about the effect of embryo integrity on the viability post-implantation, shown by the successful pregnancy rate16. Besides, other studies also used fewer blastomeres for sex diagnosis. Still, the protocol required the propagation step of cell culturing to ensure adequate material for the sexing assay. Consequently, it prolongs the test period leading to undesirable effects for embryo transfer plans, increases cost due to embryo preservation. Here, we reduced the risk of embryo damage and speeded up sexing process by taking only one blastomere and performing direct PCR for sex determination.
We used an XYClone laser system to precisely open the zona pellucida and collect the single blastomere. This technique is safer compared with using a microblade or fine needle to isolate the cell. Moreover, many studies showed that using the XY-laser system significantly improved the hatching rate of the embryos 25, 26. However, in this study, we have not validated the viabilities of the embryo post-biopsy. Therefore, further experiments are needed to confirm and optimize the blastomere isolation procedures.
To ensure the assay sensitivity even using a single cell, we removed the extraction step, which may increase the risk of loss or damage of genomic DNA, leading to the false-negative conclusion, and performed the direct PCR. Additionally, we used a robust DNA polymerase, which is more suitable for direct-PCR setting due to the resistance with various PCR inhibitors and possessing high amplification sensitivity, following claims from the manufacturer (Salagene, HCMC, Vietnam). Furthermore, we also used ammonium sulfate as a PCR enhancer, which was supposed to have a destabilizing effect, increasing the stringency during the primer-template base-pairing, thereby enhancing specificity and improving the PCR yield27. These experimental settings resulted in successfully specific amplification of BY marker and in heightening the amplified product intensity comparable to the amplified from genomic DNA, even using a very limited DNA of one single double-strand DNA copy. In another study, Park et al. reported 92.1 % accuracy of bovine sexing from single blastomere28. Using the established protocol, we succeeded in sex determination for all tested samples with 100 % sensitivity and specificity. However, due to the limitation of the embryo number and the non-random sex of the sample set, it is necessary to train and validate the method in a larger scale experiment with sufficient design factors to improve the reliability.
On the other hand, we designed the amplification using BY because it is widely used in many similar types of research 1, 29. However, several improvements need to be made to further enhance the reliability and sensitivity of the embryo single-cell sexing. These include the internal control indicating the proper amplification process. The other is increasing the number of the sex markers via multiplex PCR to reduce false-negative cases due to the imperfection of the genomic DNA right in the amplified region. In addition, there are other common markers for bovine sexing such as DDX3, amelogenin… 30, 31 that can also be combined in a single multiplex PCR for genetic enrichment of desired traits. These traits include productive, reproductive, and disease resistance which are more advantageous for embryo selection in the modern livestock industry. Our experiments also integrated two-amplicon primers specific for each sex chromosome, such as amelogenin, together with BY primer into multiplex PCR. However, the setting only generated the consistent amplification efficiency on the reactions with a higher template amount of four blastomeres or more (data not shown). For more detail, one-blastomere multiplex PCR showed the preferential amplification, in which BY amplicon was favored while X-chromosome amelogenin was diminished regardless of primer concentration adjustment. Further optimizations such as the primer set, PCR additives, cycling condition, cell processing are necessary to improve the sustainability of sex diagnosis from a single blastomere.
Conclusion
In summary, we established a procedure for rapid sex determination of bovine embryos by directly amplifying Y chromosome-specific DNA fragments from a single embryonic blastomere. We also discussed the possible improvement to increase the reliability of the single blastomere sexing and highlight the potential of the method for enrichment of the genotyping data for embryo selection, which significantly contributes to embryo research, endangered animal preservation, and the modern livestock industry.
Abbreviations
6-DMAP: 6-dimethynopyridine
FBS: Fetal bovine serum
FISH: fluorescence in situ hybridization
IVF: fertilization
mSOF: modified synthetic oviduct fluid
OCGs oocyte-cumulus-granulosa complexes
PBS: Phosphate-buffered saline
PCR: Polymerase chain reaction
SCNT: somatic nuclear transfer
TAE: Tris-acetate-EDTA
UV: ultraviolet
Authors contribution
The idea initiation, theoretical modeling, and implementation plan of the paper were made by the author Le Minh Thong; Trong Thoai Quang conducted the sexing experiment and prepared the manuscript; Truong Duy Pham prepared the embryos and somatic cell for the experiment, edited, revised, and submitted the manuscript; Nhat Thinh Nguyen and Thinh Vinh performed the somatic cell relating expreriment; Quoc Dinh Pham and Minh Chien Pham participated in prepraring of biopsy embryos and had suggestion about the manuscript; Ngoc Thuy Tram Nguyen and Van Thuan Nguyen contributed in revising the manuscript; Hai Yen Tran gave feedback on the technical support and manuscript.
Acknowledgments
This research is funded by Vietnam National University HoChiMinh City (VNU-HCM) under grant number C2021-28-04.

