Characterization of TERRA 9 repeat G4-binding RHAU peptides by Fluorescence Resonance Energy Transfer
- Ho Chi Minh City Open University, Vietnam
Abstract
Introduction: TERRA (telomeric repeat-containing RNA) is a G-quadruplex (G4) noncoding RNA that plays a part in cancer and aging. Here, we report the use of a fluorescence resonance energy transfer (FRET) approach to explore the binding mode of TERRA 9 repeat G4-binding RHAU peptides.
Methods: RHAU peptide was genetically incorporated with the fluorescent protein CFP/YFP, resulting in the generation of the fluorescent protein FRET and RHAU55-CFP/RHAU55-YFP.
Results: A strong hetero-FRET was observed when TERRA 9 repeats were added to the RHAU55-CFP/RHAU55-YFP mixture, indicating that TERRA 9 repeats could form G4 structures and simultaneously bind multiple proteins fused with the RHAU peptide.
Conclusion: This provides a useful approach for regulating the telomere elongation process by a complex of TERRA and functional proteins fused with the RHAU peptide.
INTRODUCTION
TERRA (telomeric repeat-containing RNA) is transcribed from DNA telomeres by DNA-dependent RNA polymerase II1, 2. TERRA is located in nuclear RNA fractions from eukaryotic cells such as humans, yeasts, mice and various plants. Interestingly, TERRA molecules associate with telomeric proteins and telomeres, where they are involved in the regulation processes of telomeres3. The fact that TERRA plays a crucial role along with telomerase to regulate the length of telomeres on chromosome ends correlates with cellular cancer and aging 4. Recent studies on TERRA sequences have revealed that the transcripts of C-rich single-stranded DNA telomeres are more productive than the transcripts of G-rich single-stranded DNA telomeres, leading to TERRA having G-rich sequences1, 5, 6. The G-rich TERRA contains repeats of three guanines that are able to form a four-stranded structure, known as G-quadruplex (G4)6, 7, 8. This G4 structure may be crucial for biological processes in telomeres9, 10.
G4s are four-stranded helical structures that can be formed in G-rich sequences by stacking multiple G-tetrads 11, 12, 13. G4 structures can adopt parallel and nonparallel topologies 14, 15. Analysis of the human genome demonstrated approximately 700,000 potential sequences that are able to create G4 structures16. In the genome, G4s have been found abundantly at gene promoters and telomeres17, 18. In addition, G4s also exist in the encoded RNA 5'-UTR 19 and the long TERRA8. G4 formation is involved in various cellular processes, such as transcription, translation, replication and telomere maintenance20, 21. Therefore, the G4 molecule has currently been considered a promising target for drug design in cancer treatment22, 23, 24.
Specific recognition of G4s by ligands such as small molecules, peptides and proteins provides a potential approach for the modulation of many biological processes21. Recently, RHAU protein (RNA helicase associated with AU-rich element) has been investigated that is able to bind and unwind the G4 structure specifically25, 26, 27, 28. A G4-specific binding domain (16 aa) was found in the N-terminus of the RHAU protein that specifically binds and stabilizes only parallel G4s29, 30. A study of the NMR structure solution of RHAU18 (18 aa) and a parallel DNA G4 (T95-2T) complex has shown that RHAU18 selectively binds the G-tetrad surface of G4. The α-helix structure of RHAU covers and clamps the G-tetrad surface via three-anchor-point electrostatic interactions between three positive aa of RHAU and phosphate groups of G4 (negative charge)29. Specific binding of G4 to RHAU has been applied for various biochemical applications30, 31, 32, 33, 34, 35. Fusing a fluorescent protein with the RHAU peptide generated a useful probe for distinguishing different G4 structures30. Development of a useful nuclease and ribonuclease by fusing the RHAU peptide with the fok1 (catalytic domain) and the RNase HI (catalytic domain) that can efficiently cleave the DNA and RNA in a site-specific manner, respectively34. In a previous study, TERRA 4 repeats G4-binding RHAU peptide was identified by using the electrophoresis mobility shift assay (EMSA)30. However, the type of binding mode of the TERRA 9 repeat UU(AGGGUUA)-induced self-assembly of multiple RHAU peptides has not yet been reported.
The FRET (fluorescence resonance energy transfer) technique has been used as a powerful tool to detect molecule-induced protein self-assembly or protein–protein interactions36, 37, 38. FRET signal occurrence depends on a distance range of approximately 1-10 nm between a donor molecule and an acceptor molecule39. In this report, a fluorescent protein FRET pair was genetically generated by fusing RHAU55 peptide with CFP (Cyan Fluorescent Protein) and YFP (Yellow Fluorescent Protein). TERRA 9 repeat-induced binding of both RHAU55-CFP and RHAU55-YFP can be physically detected by energy transfer from donor CFP to acceptor YFP (Figure 1). This would provide a simple approach for both characterizations of the interaction of TERRA G4 and multiple proteins in biomedical applications.

Schematic representation of TERRA 9 repeat G4-binding proteins that can be observed by the hetero-FRET method.
MATERIALS AND Methods
Construction of plasmids
A pair protein for FRET, RHAU55-CFP/RHAU55-YFP, was generated by fusing the RHAU peptide (55 aa) with CFP and YFP. DNA encoding the RHAU peptide was amplified by PCR using the gene as the template and the primer pair ON5/ON6 (ON5: 5'- gcg tgg atc cgt cca tgc atc ccg ggc acc tga aag-3"; ON6: 5'- gat tca tat ggc tgc cgc cgc cgc tct tcg ctt gaa cag aat tca gta ac-3'). The PCR product was inserted into treated pETDuet1 (Merck Millipore, Germany) at the BamHI and NdeI sites, resulting in the generation of the plasmid pRHAU. DNAs encoding YFP and CFP were amplified by PCR using pHT58430 containing the gene and pHT58240 containing the gene as templates and the primer pair ON7/ON8 (ON7: 5’-tta aag atc tag gcg gcg gca gca tgg tga gca agg gcg agg ag-3’; ON8: 5’-cca tct cga gtt act tgt aca gctcgt cca tgc cga gag tg-3’). The PCR products were then inserted into treated pRHAU at BglII and XhoI sites, resulting in the generation of plasmids pRHAU55-YFP and pRHAU55-CFP, respectively.
A plasmid for the expression of the negative control protein CFP was genetically generated by directly cloning the gene into the vector pETDuet1. DNA encoding the CFP protein was amplified by PCR using the gene as the template and the primer pair ON9/ON10 (ON9: 5'- tta agg atc caa tgg tga gca agg gcg agg ag-3'; ON10: 5'- tgc cat atg gtt act tgt aca gct cgt cca tgc c -3'. This PCR product was inserted into treated pETDuet1 at the BamHI and NdeI sites, resulting in the generation of the plasmid pCFP.
Expression and purification of proteins:
The plasmids pRHAU55-YFP (coding RHAU55-YFP), pRHAU55-CFP (coding RHAU55-CFP) and pCFP (coding CFP alone) were chemically transformed into BL21(DE3). The were cultured in Luria-Bertani medium (10 g NaCl, 5 g yeast extract and 10 g peptone in 1 L Milli-Q water) with 100 mg/L ampicillin, and the cells were incubated at 37 °C with shaking at 220 rpm to reach an OD600 of 0.8-1, IPTG (0.3 mM) (Sigma Aldrich, St. Louis, MO, USA) was subsequently added to the cell culture. The cells were continuously incubated for 12 hours at 16 °C and shaken at 180 rpm before being harvested. The pellet of cells was then resuspended in a reagent (bugBuster protein extraction, EMD Millipore, Burlington, MA, USA) consisting of bezonase nuclease (DNA and RNA degradation). The insoluble material was removed by centrifugation at 18,000 rpm for 1 hour at 4 °C. The soluble supernatant was added to the His-tag column (ThermoFisher Scientific, Waltham, MA, USA). After that, the His column was washed with 30 volumes of the column with washing buffer (20 mM Tris-HCl, 100 mM NaCl and 10 mM imidazole, pH 7). The target proteins were finally eluted with lysis buffer (20 mM Tris-HCl, 100 mM NaCl and 200 mM imidazole). The imidazole chemical in the protein solution was then removed by centrifugation using the Amicon Ultra15 (EMD Millipore). The pure target proteins were collected and analyzed by SDS–PAGE.
Circular dichroism spectroscopy
Circular dichroism (CD) measurements for the determination of the TERRA structure were performed on a spectropolarimeter (JASCO-815) using a path length quartz cuvette (10 mm) in a 40 µl reaction volume at 25 °C. Scanning from 220 nm to 320 nm was performed at a rate of 200 nm/min, pitch (1 nm), and bandwidth (2 nm). The TERRA sample was measured at concentrations of 10 µM in a buffer of 20 mM sodium phosphate and 100 mM KCl, pH 7. An average of two scans were taken for each measurement.
FRET measurements
All samples for FRET were measured in a buffer solution consisting of 20 mM potassium phosphate, pH 6.5, in quartz cuvettes with a 10 mm path length (Hellma). The sample of proteins was used at a concentration of 1 µM for the measurement. All of the FRET measurements were performed using a Cary Eclipse spectrophotometer (Varian), and all fluorescence data were automatically recorded at room temperature with a 410 nm excitation wavelength. All parameters of measurements were kept constant over the measurements to ensure all data comparison.
RESULTS
Construction of plasmids, protein expression and purification
The RHAU peptide bearing 16 amino acids (aa 53-68) is able to recognize and bind a parallel G4; however, the RHAU peptide length also influences G4 binding affinity . In this study, the fluorescent protein FRET RHAU55-YFP/RHAU55-CFP pair was generated by fusing 55 amino acids of RHAU peptide (53-107) to YFP/CFP. All DNA sequences encoding RHAU55-YFP, RHAU55-CFP and CFP were analyzed and confirmed by DNA sequencing. Proteins were expressed in the BL21 (DE3) upon the addition of IPTG. Proteins bearing a His-tag at the N-terminus were purified by chromatography using a His-tag column. The electrophoresis of proteins on SDS–PAGE showed that pure target proteins were collected (Figure 2). The molecular weights of RHAU55-YFP and RHAU55-CFP are 36,3 Da and 36,2 Da, respectively, which relocated between the bands of 30 kDa and 40 kDa of the ladder. The reference CFP is 28,6 Da, which relocated between the bands of 25 kDa and 30 kDa of the ladder.

The purified proteins were confirmed by SDS–PAGE: CFP (lane 1; 28.6 KDa), RHAU55-CFP (lane 2; 36.2 KDa) and RHAU55-YFP (lane 3; 36.3 KDa).
Circular dichroism spectroscopy
CD spectroscopy as a primary method was applied to characterize the G4 structure to evaluate the TERRA 9 repeat structure topology. TERRA RNA UU(AGGGUUA) was treated with 100 mM KCl for CD measurement. The CD spectrum of TERRA, measured at a concentration of 10 µM, displayed a positive peak at 265 nm along with a trough at 245 nm (Figure 3). This confirmed that TERRA 9 repeats formed a parallel G4 topology under K conditions.
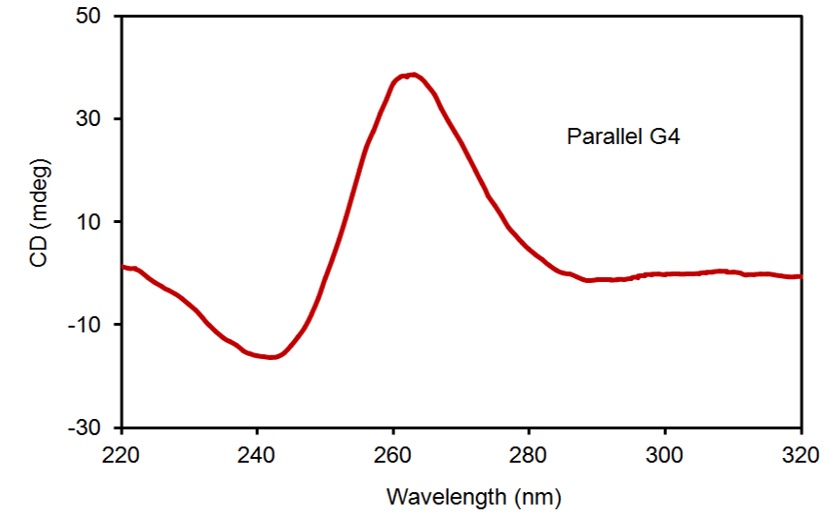
CD spectrum of TERRA forming a parallel G4 topology under K+ conditions.
Fluorescence resonance energy transfer (FRET) measurements
Herein, we used the hetero-FRET system to evaluate the binding mode of TERRA 9 repeats G4 and RHAU peptides. Incorporating RHAU peptide into a donor CFP and an acceptor YFP resulted in the construction of a pair of fluorescent protein FRET RHAU55-CFP/RHAU55-YFP, respectively. The addition of TERRA 9 repeats G4 (1 µM) to a mixture of RHAU55-CFP (1 µM) and RHAU55-YFP (1 µM) resulted in the occurrence of a strong hetero-FRET (Figure 4a). In fact, the hetero-FRET signal is obtained from 50% heterodimerization, while the remaining 50% homodimerization of protein does not donate to the hetero-FRET signal in the mixture of TERRA 9 repeats and proteins. The addition of TERRA 9 repeats G4 (1 µM) to a mixture of RHAU55-CFP (1 µM)/RHAU55-YFP (1 µM) led to an increase in the peak ratio of 527 nm/475 nm from 0.52 to 1.72 (Figure 4b). Hetero-FRET was physically observed in TERRA 9 repeat G4-dependent concentrations. The FRET signal increased with increasing concentrations of TERRA 9 repeats G4 (Figure 4a). These results showed that TERRA 9 repeats G4 could bind two RHAU peptides simultaneously. In contrast, the addition of TERRA 9 repeats G4 (1 µM) to a solution of reference CFP (1 µM) and RHAU55-YFP (1 µM) did not increase the peak ratio 527 nm/475 nm (Figure 4b). HeteroFRET was not observed in the mixture of CFP/RHAU55-YFP in the addition of TERRA, which revealed that TERRA 9 repeats G4 did not bind any side chain of CFP and YFP.
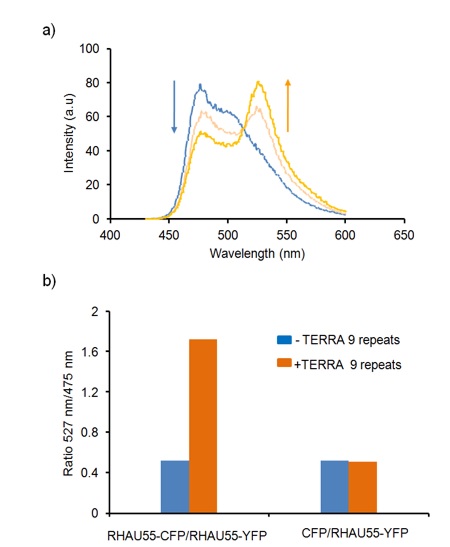
Hetero-FRET study with a pair of RHAU55-CFP/RHAU55-YFP upon the presence of TERRA 9 repeats G4. a) Representative spectra of a solution of RHAU55-CFP (1 µM)/RHAU55-YFP (1 µM) in TERRA 9 repeats at G4-dependent concentrations (0 µM, 0.5 µM and 1 µM). b) Comparison of the peak ratios at 527 nm/475 nm of FRET observed without (blue bars) and with TERRA 9 repeats (orange bars) (1 µM) for variant mixtures of proteins: RHAU55-CFP/RHAU55-YFP (both at 1 µM) and negative control CFP/RHAU55-YFP (both at 1 µM).
DISCUSSION
TERRA molecules associate with telomeric proteins and telomeres, in which they play a part in the regulation of telomere length. The formation of the G4 structure in TERRA may also be crucial for biological processes in the telomere. Characterization of the TERRA G4 structure by CD and NMR shows that TERRA remains as the parallel G4 structure topology in K+ solution7. Additionally, using a simple tool as a fluorescent protein probe specific for G4 can also determine the parallel G4 structure of TERRA30. The recognition of G4 by ligands has been of interest in the regulation of various biological processes. RHAU has been recently studied as a parallel G4-binding ligand. This study of insight into the complex of RHAU and a parallel G4 (T95-2T) demonstrates that the α-helix of RHAU peptide selectively clamps on the surface of the G-tetrad by 3-anchor-point electrostatic interactions. However, the binding mode of the complex of RHAU and TERRA 9 repeats G4 has not yet been characterized. Thus, the development of an alternative and simple FRET method for the detection of the binding mode of TERRA 9 repeats G4 and RHAU peptide needs to be established. A strong hetero-FRET signal from donor (CFP) to acceptor (YFP) was observed in the mixture of RHAU55-CFP/RHAU55-YFP under the addition of TERRA 9 repeats G4, which demonstrates that TERRA 9 repeats G4 can specifically bind multiple RHAU peptides simultaneously. In addition, titration of TERRA 9 repeats G4 to the mixture of RHAU55-CFP and RHAU55-YFP showed that the FRET signal increased with increasing concentrations of TERRA 9 repeats G4, which may provide a potential device for detecting the level of TERRA expression in cancer cells.
CONCLUSIONS
TERRA 9 repeats RNA is capable of forming a parallel G4 that can bind multiple RHAU peptides simultaneously. The fluorescent protein FRET pair was constructed by fusing the RHAU peptide with CFP and YFP, allowing for detecting the interaction of TERRA G4 and RHAU peptides. This opens up a new approach for the characterization of TERRA 9 repeats G4 in which TERRA can recruit multiple self-assemblies of functional proteins fused with RHAU peptide for biomedical applications.
Abbreaviations
CD: Circular dichroismCFP: Cyan fluorescent proteinDNA: Deoxyribonucleic acidEMSA: Electrophoresis mobility shitf assay FRET: Fluorescence resonance energy transferG4: G-quadruplexNMR: Nuclear magnetic resonancePCR: Polymerase chain reactionRNA: Ribonucleic acidRHAU: RNA helicase associated with AU-rich elementTERRA: Telomeric repeat-containing RNAUTR: Untranslated regionYFP: Yellow fluorescent protein
Acknowledgement
We would like to thank Prof. Anh Tuân Phan (Nanyang Technological University, Singapore) for scientific discussion and kindly giving plasmids.
COMPETING INTERESTS
There are no conflicts of interest.

