Identification of commensal Pseudomonas aeruginosa isolates using duplex PCR targeting the oprL and algD genes
- School of Biotechnology, International University, Research Center for Infectious Diseases, International University, Vietnam National University of HCMC
- School of Biotechnology, International University
- Vietnam National University of HCMC
Abstract
Background: Pseudomonas aeruginosa (P. aeruginosa) is a ubiquitous bacterium which can be found in most moisture places such as soil, water, food, plants, and animals including humans. Due to genetic flexibility among strains, there is no standard molecular identification for P. aeruginosa from different sources, particularly for human colonizing P. aeruginosa or commensal isolates.
Materials and Method: Monoplex PCR using published primers of oprL, algD, nfxB gene were compared for their specificity and sensitivity in identification of commensal P. aeruginosa. Twenty- nine human colonizing Pseudomonas isolates including 16 P. aeruginosa isolates and 13 P. aeruginosa-like isolates were used in the study. To improve the detection, two new primer pairs targeting oprL gene was designed (oprL-pp1 and oprL-pp2), and oprL-algD duplex PCR was carried out with newly designed oprL primer pairs.
Result and conclusion: PCR targeting algD or nfxB genes has the same sensitivity of 93.75% and specificity of 100%, while oprL-specific PCR using published primers is more sensitive (100%) but less specific (0%). Duplex PCR yielded high sensitivity and specificity in detecting P. aeruginosa. Both oprL-pp1/algD and oprL-pp2/algD duplex-PCR had 93.75% sensitivity (15/16 P. aeruginosa isolates) and 100% specificity (0/13 P. aeruginosa-like isolates). Besides, oprL-pp2 primers were more specific than oprL-pp1 primers, with only 2 over 13 P. aeruginosa-like isolates detected, while oprL-pp1 primers detected all P. aeruginosa-like isolates. Compared to the monoplex PCR that only targeted oprL gene, the duplex-PCR utilizing oprL-pp2 and algD primer can identify 15/16 P. aeruginosa isolates (93.75% specificity) and reduce the number of oprL-positive isolates required further exact identification. Additionally, the duplex-PCR used in this study was negative for non-Pseudomonas species including E. coli, V. cholera, V. parahaemolyticus, S. aureus. In conclusion, our duplex PCR targeting oprL and algD could be a valuable tool for P. aeruginosa screening.
Introduction
is a ubiquitous bacterium that can be found in water, soil, food, plants and animals. It can also be isolated from healthy people’s skin, throat, and stool and is considered part of the human commensal flora; thus, it is called commensal bacteria 1. As a serious life-threatening Gram-negative pathogen, they are responsible for a wide range of minor to severe infections in burn patients, immunocompromised individuals or patients with cystic fibrosis. These infections are difficult to eliminate due to the pathogen’s intrinsic and extrinsic resistance abilities. Therefore, early and accurate detection of is critical for treating infected patients.
Many detection methods have been developed, including traditional bacterial culture methods, immunological assays, and biochemical tests 2. However, these tests typically take days to weeks to complete confirmation, with a high rate of misidentification due to cross reactions of with other related Gram-negative bacilli. In recent years, polymerase chain reaction (PCR) has been developed as a rapid and reliable molecular method for clinical and environmental detection using a number of specific genes, such as . 3, 4, 5 Multiplex PCR, which allows multiple target detection in a single PCR run, was also considered for clinical 6. Commensal isolates, on the other hand, have been investigated even though the detection of these strains is important for tracking and analysis of the pathogen and the spread of antibiotic resistance status 7. In this study, the application of PCR and multiplex PCR for commensal isolates was investigated.
Materials and Methods
Bacterial isolates
Twenty-nine commensal isolates were used in this study. They included 16 isolates and 11 , 1 and 1 isolates, which were identified via 16S rRNA sequencing (Nam Khoa Biotek CO., Ltd).
DNA extraction
Bacterial DNA was extracted by the organic phenol‒chloroform method 8 and quantitated using Take3 microvolume plates (Synergy HT, Biotek). For long-term storage, DNA was stored in 1X TE (Tris-EDTA) buffer at -20°C.
Primer design
Two primer sets, pp1 and pp2 (
Monoplex and duplex PCR for the detection of
PCR was carried out in a total reaction volume of 25 µL containing 12.5 µL master mix (iStandard iVAPCR 2X Master Mix, Viet A Technology Corporation, Vietnam), 2 µL DNA template and either 0.5 µM of primers, 0.4 µM primers or 0.4 µM primers (monoplex) or in combination of 0.4 µM of each and primer (duplex) (PHUSA Biochem Ltd., Vietnam). PCR was carried out as described in
Data analysis
The sensitivity and specificity were determined by the formula from Rajul Parikj MS 9. In short, sensitivity was calculated as true positive/(true positive+ false negative), and specificity was calculated as true negative/(true negative+ false positive). The data were analyzed using Excel (Microsoft Office, USA).
Results
Monoplex PCR using -pp0 detected all commensal isolates but not specifically

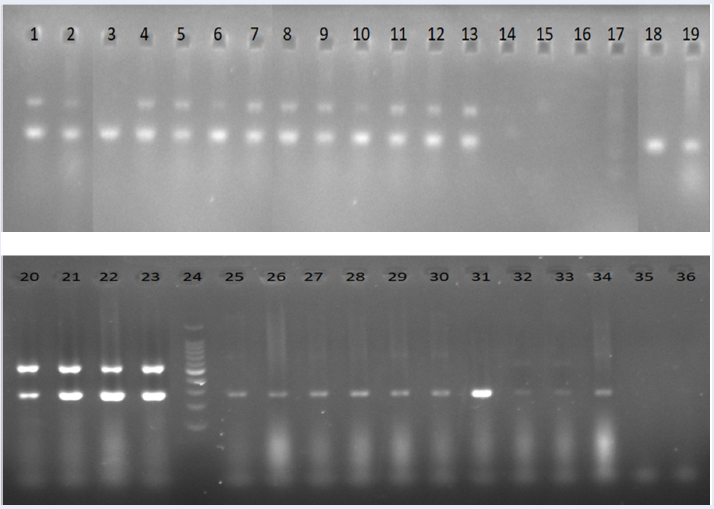
Representative PCR products of the oprL gene (oprL-pp0; 504 bp) in Pseudomonas aeruginosa. Lane 1: 100 bp DNA ladder; lane 2: P. aeruginosa ATCC 9027; lane 3: Negative control; lane 4 – 11: samples with positive results; lane 12: sample with negative results.
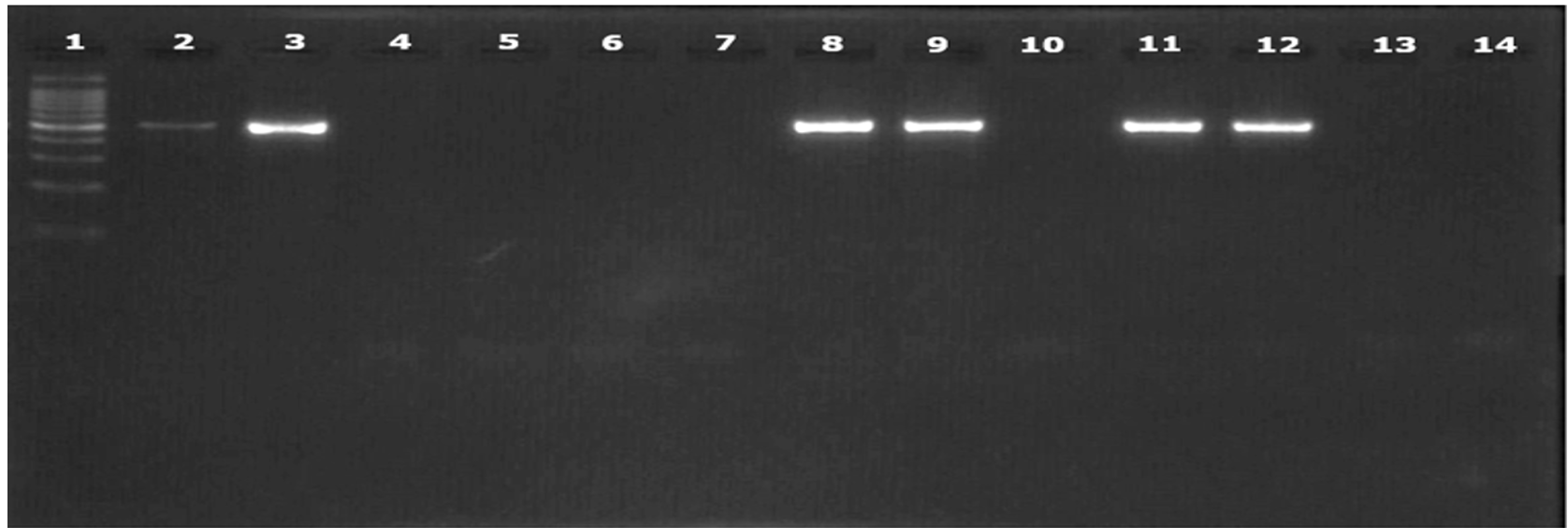
Representative PCR products of

Representative PCR products of

Duplex PCR using
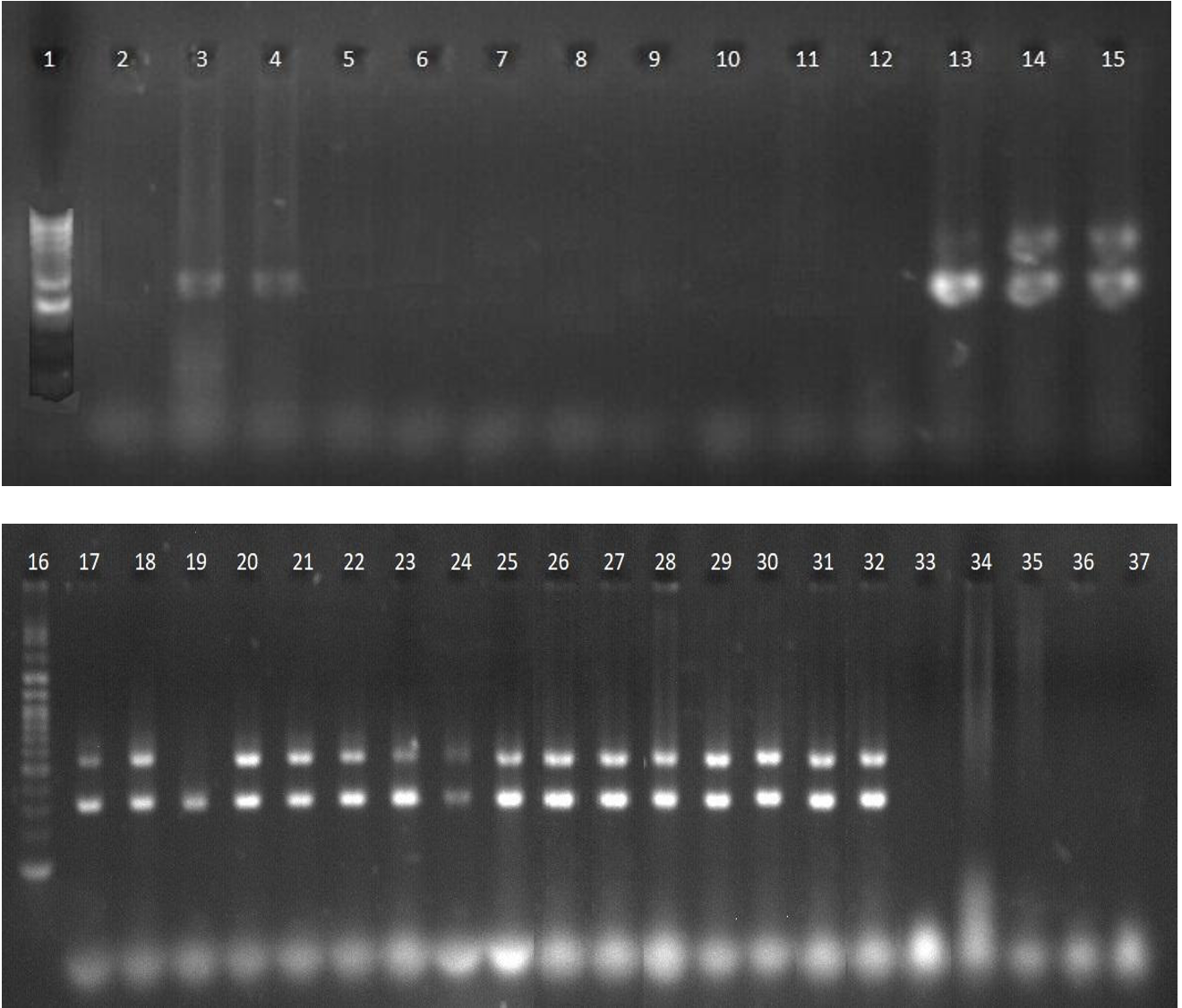
Duplex PCR using
PCR using -pp0 primer pairs detected all 16 isolates; thus, the sensitivity was 100%. However, it failed to differentiate isolates from other species, including , and , resulting in a specificity of 0% (Figure 1,
Characteristics of published and designed primers and nucleotide sequences
|
Primers |
Oligonucleotide sequence (5’ to 3’) |
PCR program |
Target |
Primer Length (bp) |
Amplicon Size (bp) |
Reference |
|
oprL-pp0-F oprL-pp0-R |
5’-ATGGAAATGCTGAAATTCGGC-3’ 5’-CTTCTTCAGCTCGACGCGACG-3’ |
30 cycles: 2 min 95oC, 30 s 56oC, 1 min 72oC |
oprL gene |
21 |
504 |
|
|
algD-F algD-R |
5’ TTCCCTCGCAGAGAAAACATC 3’ 5’ CCTGGTTGATCAGGTCGATCT 3’ |
30 cycles: 1 min 94oC, 1 min 60oC, 1 min 72oC |
algD gene |
21 |
520 |
|
|
nfxB-F nfxB-R |
5’-CGATCCTTCCTATTGCACGC-3’ 5’-AGGGTGATGAACAGTTCGGT-3’ |
30 cycles: 1 min 94oC, 45 s 49.3oC, 2 min 72oC |
nfxB gene |
20 |
673 |
|
|
oprL-pp1-F oprL-pp1-R |
5’-GCGATCACCACCTTCTACTT-3’ 5’-CGGACGCTCTTTACCATAGG-3’ |
30 cycles: 1 min 95oC, 1 min 60oC, 1 min 72oC |
oprL gene |
20 |
258 |
This study |
|
oprL-pp2-F oprL-pp2-R |
5’-CTGTGGGTTGCTCCTCCAAG-3’ 5’-CTCGCCCAGAGCCATATTGT-3’ |
30 cycles: 1 min 95oC, 1 min 60oC, 1 min 72oC |
oprL gene |
20 |
299 |
This study |
Specificity and sensitivity of
|
PCR |
Duplex |
Duplex |
Monoplex | ||
|
Primer |
|
|
|
|
|
|
Confirmed P. |
16/16 |
15/16 |
16/16 |
15/16 |
16/16 |
|
Liked P. |
2/13 |
0/13 |
13/13 |
0/13 |
13/13 |
|
E. |
0/1 |
0/1 |
0/1 |
0/1 |
0/1 |
|
S. |
0/1 |
0/1 |
0/1 |
0/1 |
0/1 |
|
V. |
0/1 |
0/1 |
0/1 |
0/1 |
0/1 |
|
V. |
0/1 |
0/1 |
0/1 |
0/1 |
0/1 |
|
Sensitivity |
100% |
93.75% |
100% |
93.75% |
100% |
|
Specificity |
84.62% |
100% |
0% |
100% |
0% |
Monoplex PCR targeting and detected specifically
PCR using and PCR using gave identical results.All non- samples were negative. All isolates gave positive results except one isolate (isolate 234.1) (Figure 2, Figure 3). The specificity of PCR using and for detecting was 100%, while the sensitivity was 93.75%.
Duplex PCR targeting and
Two primer pairs for were designed in this study with the aim of maintaining the ability to detect 234.1 but not other non-PA isolates. They were coupled with in a duplex PCR to design an effective identification method for commensal .
The results showed that pp1 still detected 13/13 non-isolates, while pp2 could differentiate some of the non-isolates. In addition, both primer pairs in the duplex were negative for all other bacterial species ( (
Both the-pp2/ and-pp1/ duplex PCR tests had the same detection efficiency, with 100% specificity and 93.75% sensitivity, since there was one isolate (isolate 234.1) detected by primers but not by (Figure 4, Figure 5). The isolate 234.1 showed only one band for in the electrophoresis gel in both duplex PCR assays.
More importantly, duplex PCR using pp2 primers, while maintaining 100% sensitivity to detect , such as PCR using -pp0 and pp1 primer pairs, has a specificity of 84.62%, as only 2/13 -like isolates gave positive results (
Discussion
Many studies have found that the PCR approach has a higher sensitivity than the classic culture approach in detecting , especially at the beginning of colonization10, 11. Along with the advancement of PCR tests for , a number of exclusive genes have been found. Khan and Cerniglia developed the first PCR technique for detecting based on the exotoxin A gene 11. , and genes and other genes were also considered in monoplex or multiplex PCR tests for identification 4, 12, 13.
and are two exclusive genes that have been used in monoplex PCR to detect with high specificity and sensitivity. While the sensitivity of both genes was greater than 90%, the specificity of was 100% and only slightly higher than 80% for . encodes for GDP — mannose 6 — dehydrogenase of the alginate synthesis pathway 14, while the o gene encodes for an outer membrane protein that plays important roles in the interaction of this pathogen with the environment 15. a repressor of the MexCD-oprJ efflux pump, is another potential target for detection 16, 17. In this study, the monoplex results showed that maintained the specificity of but did not improve sensitivity. Both monoplex PCRs targeting the and genes failed to detect 100% of the isolates. PCR targeting only a single fragment of the gene results in a lack of precision, as clinical strains display high genotypic variability 18. In fact, several studies have reported the absence of one or more virulence genes in certain strains of
Multiplex PCR is more efficient than monoplex due to its ability to simultaneously amplify multiple PCR products in a single reaction, allowing for multiplex detection and greatly reducing the cost and time requirements. Aghamollaei et al. developed a detection assay using triplex PCR that amplifies the , and genes, which successfully identified 100% of the clinical isolates tested 6. However, the specificity of this multiplex PCR against -like isolates was not fully considered. Another PCR assay using the same genes was effective in identifying 95% of isolates from Dorper sheep milk 21. Multiplex PCR can also be developed to allow for more in-depth diagnostics, as multiple bands and single bands can be interpreted differently, minimizing false positives and false negatives 22.
This study considers the possibility of a duplex PCR detection method using coupled with or genes. PCR targeting the or genes has a sensitivity of 93.75% and specificity of 100%, while is more sensitive but less specific. Although the De Vos study demonstrated that the published -pp0 primers could sufficiently detect from other species8, our data suggested that the primer pairs also detect non- species. In a previous report, the same -specific primer set also had just 70% specificity, with only 49 out of 70 -positive clinical samples being . Improvement of the specificity of the primers might improve the specificity of this PCR assay. Duplex PCR of resulted in 2 bands, indicating confirmed isolates, while one band indicated that further identification methods were needed to confirm the isolate identity, and no band was non-. At the same time, by designing primers with the same annealing temperature as or , we can develop a cost-effective duplex PCR to rapidly detect
The designed -pp2 primer pair in this study was more specific in detection than -pp1 and pp0, with only 2 false-positive results (2/13), while the -pp1 primer pair gave positive results for 13/13 -like isolates (
-pp2 duplex PCR is a simple, specific, and sensitive method for identification. It is a cost-effective and time-saving method because the processing time from sample preparation to confirmation completion is less than 3 hours. Although L-pp1 and -pp2/ duplex PCR have the same sensitivity and specificity, the latter produced more precise results since non- samples were less likely to be positive The 2-target system of the assay decreases the potential for sequence-related false negatives and can provide simultaneous confirmation of positive results.
Conclusion
In conclusion, the results of this study showed that, using the newly designed primers, the duplex PCR assay targeting the and genes was able to identify with improved specificity. Although additional confirmation of the accuracy of this approach is needed, the results imply that the PCR test presented in this article is a simple, rapid and sensitive tool for the early identification of commensal isolates.
List of abbreviations
:
PCR: Polymerase chain reaction
TE: Tris-EDTA
SB: Sodium borate
COMPETING INTERESTS
The authors declare that they have no competing interests.
Acknowledgments
This research is funded by Vietnam National University of Ho Chi Minh City under grant number C2020-28-03.
Author’s contributions
Thuc Quyen Huynh wrote the first draft; Nguyen Bao Vy Tran performed the experiments; Thi Thuy Vy Pham collected the isolates; Nguyen Huong Giang Vo performed the experiments; Lam Que Anh Nguyen collected the isolates; Ngoc My Huong Nguyen collected the isolates; Van Dung Nguyen performed the experiment; Thi Thu Hoai Nguyen designed, supervised and reviewed the work.

