Bacterial pathogens causing community–acquired pneumonia in hospitalized adult patients with and without chronic obstructive pulmonary disease
- University of Medicine and Pharmacy HCMC
- Nhan dan Gia Dinh Hospital
- Nam Khoa Biotek
Abstract
Background: Community-acquired pneumonia (CAP) is a common infection that often occurs in older adults who may have chronic obstructive pulmonary disease (COPD), a common respiratory condition characterized by airflow limitation. Chronic obstructive pulmonary disease and community-acquired pneumonia usually cause the same symptoms as respiratory tract infections, but they have potential differences in microbial etiology. This study aimed to assess the potential of bacterial pathogens in hospitalized CAP patients with and without COPD as well as bacterial combinations and to examine different rates of bacterial pathogens causing CAP between patients with and without COPD.
Methods: This is a multicenter study conducted on hospitalized adult patients with community-acquired pneumonia with and without chronic obstructive pulmonary disease at the Respiratory Department of Nguyen Tri Phuong Hospital, Nhan Dan Gia Dinh Hospital and University Medical Center from 04/2021 to 03/2023. Collected sputum samples that were assessed as reliable (according to the Bartlett scale) were included in the study. The sputum samples were transported to Nam Khoa Company’s laboratory to carry out multiplex real-time PCR with King Fisher FLEX as the nucleic acid extraction instrument and CFX 96TM of Bio-Rad as the real-time PCR system. For statistical analysis, data collection was solved by SPSS 20.0 software and Microsoft Excel 2020.
Results: Among 341 CAP patients, there were 91 patients (26.7%) with COPD, in which 89 patients were detected with a bacterial infection. The positive rates were 97.8% in CAP patients with COPD and 54.0% in CAP patients without COPD (p<0.001). Bacterial pathogens that caused CAP in patients with and without COPD extended to gram-negative bacilli. The top 5 bacterial pathogens in CAP patients with and without COPD were Acinetobacter baumannii (25.3% & 14.4%), Haemophilus influenzae (23.1% & 10.8%), Klebsiella pneumoniae (22.0% & 17.2%), Streptococcus pneumoniae (20.9% & 14.8%) and Escherichia coli (13.2% & 8.4%), in which the different percentages of Acinetobacter baumannii and Haemophilus influenzae were statistically significant (p<0.05). Pseudomonas aeruginosa was found at a low frequency (1.1% & 5.6%). Atypical bacteria were detected for only Mycoplasma at low frequencies (4.4% & 6.8%) and often occurred as a combined bacterium. Klebsiella pneumoniae and Escherichia coli in CAP with COPD and Acinetobacter baumannii and Escherichia coli in CAP without COPD were not often defined as the primary bacteria alone. More than one bacterial pathogen was commonly found in the sputum of CAP patients with and without COPD.
Conclusions: CAP patients with COPD occur at a rate of 26.7%. Bacterial pathogens were detected in 97.8% of CAP patients with COPD and 54.0% of CAP patients without COPD (p<0.001), and they extended to gram-negative bacilli. The top 5 bacterial pathogens in the two groups were the same with different rates, in which the different rates of Acinetobacter baumannii and Haemophilus influenzae were statistically significant (p<0,05). Pseudomonas aeruginosa is found less commonly, although it is important because of its critical antibiotic resistance and mortality. Atypical bacteria are detected for only Mycoplasma in low frequency, and it often occurs as a combined bacterium. Klebsiella pneumoniae and Escherichia coli in CAP with and without COPD are rarely or not defined as the primary bacteria alone. More than one bacterial pathogen is commonly found in the sputum of CAP patients with and without COPD.
INTRODUCTION
Community-acquired pneumonia (CAP) is a common infection that occurs in any individual at any age, especially in older adults, who may have chronic obstructive pulmonary disease (COPD), a common respiratory condition characterized by abnormalities of the airways1. Chronic obstructive pulmonary disease is also one of the leading causes of morbidity and mortality worldwide. Recent projections predict that by 2030, it will be the fourth main cause of death and the seventh cause of the global burden of infectious disease2. Chronic obstructive pulmonary disease and community-acquired pneumonia usually cause the same symptoms as respiratory tract infections, but they have potential differences in microbial etiology1, 3. Otherwise, as a consequence of its chronicity, chronic obstructive pulmonary disease causes high resource utilization with clinician office visits and frequent hospitalization due to acute exacerbation for treatment, especially with antibiotics for microbial infection4, 5, 6. Hospitalized community-acquired pneumonia, with or without chronic obstructive pulmonary disease, is hard to identify the pathogenic bacteria since the primary specimen is often taken from patients’ sputum, which is at high risk of contamination when passing through the oropharynx. Therefore, traditional cultural techniques are limited due to many difficulties7: most patients have taken antibiotics, and the bacteria could still be alive in the alveola or bronchial epithelia fluid but could have already perished in the sputum. In addition, several subjective reasons from the laboratory could reduce the ability to successfully culture pathogens, such as a lack of adequate environment to isolate the primary pathogens while they are often difficult to culture, samples that cannot be cultured as quickly as possible to increase the chance of detecting pathogens, inexperienced technicians hence unable to choose the right pathogen colonies on the isolating agar plates, or the sample was not assessed for its reliability to remove nonsputum fluid such as viscous mucus in the oropharynx prior to the isolation procedure. To overcome such difficulties, multiplex real-time PCR was used as the optimal technique, which has also proven its high sensitivity and specificity. Multiplex real-time PCR can not only simultaneously detect the specific nucleic acid sequence of the bacteria but also calculate the number of copies to allow the identification of bacterial agents as pathogens.
The aims of this study were (1) to assess the potential of bacterial pathogens in hospitalized patients with community-acquired pneumonia with and without chronic obstructive pulmonary disease as well as bacterial combinations. (2) To examine different rates of bacterial pathogens that caused community-acquired pneumonia between patients with and without chronic obstructive pulmonary disease.
METHODS
This is a multicenter study conducted on hospitalized adult patients with community-acquired pneumonia with and without chronic obstructive pulmonary disease at the Respiratory Department of Nguyen Tri Phuong Hospital, Nhan Dan Gia Dinh Hospital and University Medical Center from 04/2021 to 03/2023. Collected sputum samples that were assessed as reliable (according to the Bartlett scale) were included in the study. The samples were transported to Nam Khoa Company’s laboratory to carry out multiplex real-time PCR in which DNARNAprep-MAGBEAD (from Nam Khoa Co.) and King Fisher FLEX (from Thermo) were used for the nucleic acid extraction step; multiplex real-time PCR mixes specific for bacterial pathogens causing pneumonia (Nam Khoa Co.) and the CFX 96 real-time PCR machine (from Bio-Rad) were used for the amplification step to detect and quantify the target nucleic acid by real-time PCR. Bacterial agents were identified as pathogens when their quantitative measurement was ≥ 100.000 copies. For atypical bacteria, the detected bacteria were identified as pathogenic agents regardless of their quantity. Bacteria with the highest quantity were considered primary pathogens, and others with lower quantities were considered combined agents7, 8. For statistical analysis, data collection was performed using SPSS 20.0 and Microsoft Excel 2020 software.
For ethical considerations, we only practiced in the laboratory to detect bacterial pathogens causing community-acquired pneumonia due to the requirement of clinical doctors. The researcher had no contact with patients or clinical doctors for requirements. Our research was approved by the Independent Ethical Committee (IEC) of the University of Medicine and Pharmacy HCMC at Decision No 330/DHYD-HDDD, issue: June 14, 2019.
RESULTS
There were 341 sputum samples from hospitalized adult patients with community-acquired pneumonia that matched the inclusion criteria of this study. The demographic data and the results of bacterial detection by multiplex real-time PCR are shown in
Demographic data and bacterial detection by MPL-rPCR
|
Characteristics |
Patients with COPD (91) n (%) |
Patients without COPD (250) n (%) |
p value |
|
Gender Female Male |
9 (9.9) 82 (90.1) |
119 (47.6) 131 (52.4) |
p < 0.001 |
|
Age 16 - 60 years > 60 years |
20 (22.0) 71 (78.0) |
72 (28.8) 178 (71.2) |
p = 0.209 |
|
CAP patients |
91 (26.7) |
250 (73.3) |
p < 0.001 |
|
Positive rate |
89 (97.8) |
135 (54.0) |
p < 0.001 |
In 91 CAP patients with COPD, 89 patients were detected with bacterial pathogens. The positive rate was 97.8%. The bacterial pathogens causing CAP in 91 patients with COPD detected by multiplex real-time PCR are shown in
The proportion of bacterial pathogens in CAP patients with COPD
|
Pathogens |
N |
%* | ||
|
gram-positive (38) |
Streptococcus pneumoniae |
19 |
20.9 | |
|
Streptococcus agalactiae |
2 |
2.2 | ||
|
Staphylococcus aureus (MRSA) |
1 |
1.1 | ||
|
Staphylococcus epidermidis (MRSE) |
11 |
12.1 | ||
|
Coagulase negative staphylococcus |
2 |
2.2 | ||
|
Enterococcus faecalis |
2 |
2.2 | ||
|
Enterococcus faecium |
1 |
1.1 | ||
|
gram-negative (98) |
Enterobacteriaceae (37) |
Escherichia coli |
12 |
13.2 |
|
Klebsiella pneumoniae |
20 |
22.0 | ||
|
Morganella morganii |
3 |
3.3 | ||
|
Providencia sp. |
2 |
2.2 | ||
|
Others (61) |
Acinetobacter baumannii |
23 |
25.3 | |
|
Burkholderia cepacia |
3 |
3.3 | ||
|
Pseudomonas aeruginosa |
1 |
1.1 | ||
|
Moraxella catarrhalis |
3 |
3.3 | ||
|
Haemophilus influenzae |
21 |
23.1 | ||
|
Haemophilus influenzae type B |
1 |
1.1 | ||
|
Stenotrophomonas maltophilia |
9 |
9.9 | ||
|
Atypical bacteria (4) |
Mycoplasma sp. |
4 |
4.4 | |
The data from
Among 250 CAP patients without COPD, 135 patients were detected with bacterial pathogens. The positive rate was 54.0%. The bacterial pathogens detected by multiplex real-time PCR of the sputum samples collected from 250 CAP patients without COPD are shown in
The proportion of bacterial pathogens in CAP patients without COPD
|
Pathogens |
|
|
N |
%* |
|
gram-positive (70) |
Streptococcus pneumoniae |
37 |
14.8 | |
|
Staphylococcus aureus (MRSA) |
7 |
2.8 | ||
|
Staphylococcus aureus (MSSA) |
1 |
0.4 | ||
|
Coagulase negative staphylococcus |
2 |
0.8 | ||
|
Staphylococcus epidermidis (MRSE) |
10 |
4.0 | ||
|
Enterococcus faecalis |
5 |
2.0 | ||
|
Enterococcus faecium |
8 |
3.2 | ||
|
gram-negative (192) |
Enterobacteriaceae (88) |
Escherichia coli |
21 |
8.4 |
|
Klebsiella pneumoniae |
43 |
17.2 | ||
|
Enterobacter cloaceae |
1 |
0.4 | ||
|
Morganella morganii |
9 |
3.6 | ||
|
Providencia sp. |
9 |
3.6 | ||
|
Proteus mirabilis |
5 |
2.0 | ||
|
Others (104) |
Acinetobacter baumannii |
36 |
14.4 | |
|
Burkholderia cepacia |
6 |
2.4 | ||
|
Pseudomonas aeruginosa |
14 |
5.6 | ||
|
Moraxella catarrhalis |
1 |
0.4 | ||
|
Haemophilus influenzae |
27 |
10.8 | ||
|
Stenotrophomonas maltophilia |
20 |
8.0 | ||
|
Atypical bacteria (17) |
Mycoplasma sp. |
17 |
6.8 | |
According to
From the list of bacterial pathogens that caused CAP in patients with and without COPD, we selected the top 5 bacterial pathogens, which are shown in
The top 5 bacterial pathogens causing CAP with or without COPD
|
CAP with COPD |
CAP without COPD |
p - value | ||
|
Pathogens |
n (%) |
Pathogens |
n (%) |
|
|
Acinetobacter baumannii |
23 (25.3) |
Acinetobacter baumannii |
36 (14.4) |
0.018 |
|
Haemophilus influenzae |
21 (23.1) |
Haemophilus influenzae |
27 (10.8) |
0.003 |
|
Klebsiellla pneumoniae |
20 (22.0) |
Klebsiellla pneumoniae |
43 (17.2) |
0.314 |
|
Streptococcus pneumoniae |
19 (20.9) |
Streptococcus pneumoniae |
37 (14.8) |
0.180 |
|
Escherichia coli |
12 (13.2) |
Escherichia coli |
21 (8.4) |
0.186 |
Data from
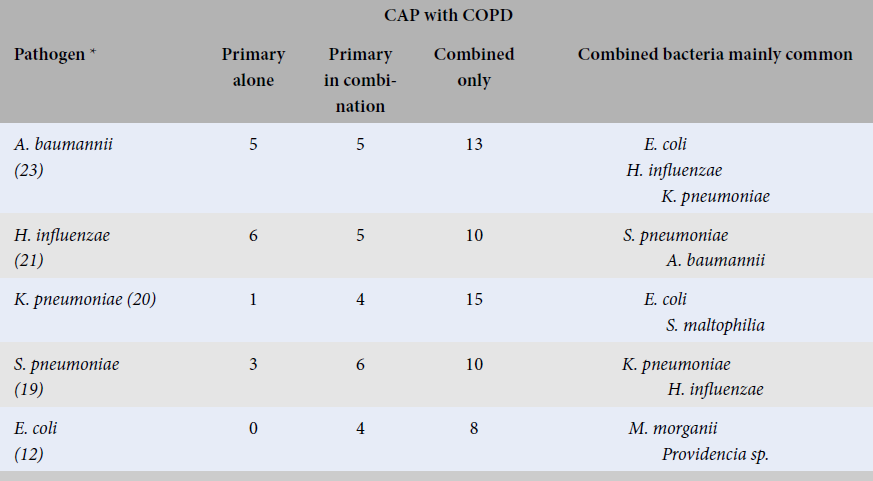
The combination of the top 5 bacterial pathogens causing CAP with and without COPD
|
CAP with COPD | ||||
|
Pathogen * |
Primary alone |
Primary in combination |
Combined only |
Combined bacteria mainly common |
|
A. baumannii (23) |
5 |
5 |
13 |
E. coli H. influenzae K. pneumoniae |
|
H. influenzae (21) |
6 |
5 |
10 |
S. pneumoniae A. baumannii |
|
K. pneumoniae (20) |
1 |
4 |
15 |
E. coli S. maltophilia |
|
S. pneumoniae (19) |
3 |
6 |
10 |
K. pneumoniae H. influenzae |
|
E. coli (12) |
0 |
4 |
8 |
M. morganii Providencia sp. |
|
CAP without COPD | ||||
|
K. pneumoniae (43) |
8 |
12 |
23 |
A. baumannii P. aeruginosa E. coli |
|
S. pneumoniae (37) |
14 |
12 |
11 |
K. pneumoniae H. influenzae M. catarrhalis |
|
A. baumannii (36) |
5 |
8 |
23 |
K. pneumoniae S. pneumoniae E. faecalis E. faecium |
|
H. influenzae (27) |
13 |
5 |
9 |
S. pneumoniae A. baumannii Mycoplasma sp. |
|
E. coli (21) |
2 |
4 |
15 |
A. baumannii M. catarrhalis |
The data from
DISCUSSION
In our study, the rate of CAP patients with COPD aged over 60 years was 78.0%, and the rate of male sex was 90.1%. These rates were similar to the reports by previous authors: Julio A., Ramirez and Rodrigo Cavallazzi (74.0% and 90.5%)9, Xue-Jun Li MD (75.3% and 71.0%)10, Joan Gómez-Junyent (79.9% and 90.5%)11, Dang Quynh Giao Vu, Le Thuong Vu (87.6% and 88.5%)12. There has been a significant increase in the age of CAP patients with and without COPD over the past decade, probably because of the increasing age of the population13. Community-acquired pneumonia patients with COPD had a prevalence rate of 26.7% (91/341), similar to the reports by Sogaard M (33.3%)14, Amir Sharafkhaneh (32.7%)15, Joan Gómez-Junyent (23.9%)11 and Pascual-Guardia (21%)16.
Although COPD was only 26.7% in total CAP patients, the proportion of bacterial pathogens (positive rate) was 97.8%, while in CAP patients without COPD, the proportion was 54.0%. There was a statistically significant difference in the pathogen detection rate between these two groups of patients (p < 0.001). The pathogen detection rate in CAP patients with COPD by Ly Khanh Van and Pham Hung Van was 69.0%17, lower than that in our research (97.8%). The pathogen detection rates in CAP patients without COPD were 53% by Xue-Jun Li MD10 and 53.8% by Dao Thi My Ha18, similar to our study (54.0%). Some previous reports indicated that the microbial etiology of CAP in patients with COPD may differ from that of CAP patients without COPD, and more than one bacterial pathogen was found in the sputum of CAP patients with and without COPD11, 13, 19, 20, 21.
In CAP patients with COPD, was the most prevalent at 25.3%, while was only at 20.9% (
In our study, the top 5 bacterial pathogens in CAP with and without COPD were the same as , , and but at different prevalence rates, in which the different percentages of and were statistically significant (p < 0.005), similar to the report by previous authors3, 10, 11, 23.
In this study, causing CAP in patients with and without COPD was counted at rates of 1.1% and 5.6%, respectively (Tables 2, 3). Although was counted at a low rate, it was important because of risk factors such as antibiotic resistance, mortality and outcomes10, 13, 17, 27, 28, 29, 30, 31. Furthermore, previous studies have reported that a played an important role in CAP patients with severe COPD who were elderly and associated with regular oral corticosteroid therapy31, 32, 33.
Atypical bacteria were detected for only at low frequency in CAP patients with COPD (4.4%) and without COPD (6.8%), similar to a previous report by De-Shun Liu (6.5%)3. Some recent studies have suggested that atypical bacteria in CAP patients are rare and often occur as combined bacteria, along with typical bacterial pathogens34, 35, 36.
In the bacterial combination, our study showed that , , , and can play a role as primary bacterial pathogens as well as combined bacterial pathogens in which and were rarely or not found as the primary bacteria alone in CAP with and without COPD.
CONCLUSIONS
Among 341 CAP patients, 91 patients had COPD (26.7%), 97.8% had bacterial pathogens detected by multiplex real-time PCR, and the positive rate in CAP patients without COPD was 54.0% (p < 0.001). Bacterial pathogens causing CAP in patients with and without COPD extend to gram-negative bacilli. The top 5 bacterial pathogens in the two groups were the same with different rates, in which the different rates of and were statistically significant (p < 0.05). is less common, although it is important because of its critical antibiotic resistance and mortality. Atypical bacteria are detected for only at low frequency and often occur as a combined bacterium. and in CAP with and without COPD are rarely or not defined as the primary bacteria alone. More than one bacterial pathogen is commonly found in the sputum of CAP patients with and without COPD.
Abbreviations
CAP: Community-acquired pneumonia
CMV: Cytomegalo virus
COPD: Chronic Obstructive Pulmonary Disease
EBV: Epstein-Barr virus
IEC: Independent Ethics Committee
MRSA: Methicillin-Resistant
MRSE: Methicillin-resistant
MSSA: Methicillin-susceptible
ACKNOWLEDGEMENT
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Conceptualization, writing original protocol: L.K.V and L.V.X.; investigation writing – original draft, and collecting data: L.K.V.; protocol review: L.V.X.; formal analysis: L.K.V; writing – review and editing: L.K.V, L.V.X and P.H.V; All authors, including L.K.V, L.V.X and P.H.V revised the manuscript and agreed to the final version before submission.

