Isolation of cellulose-degrading bacteria from sugarcane bagasse
- Department of Biotransformation Technology, Institute of Tropical Biology, Vietnam Academy of Science and Technology
Abstract
Introduction: Bagasse in the sugar industry contains a large amount of nutrients and is a very good source of organic fertilizer for plants. However, the main component of sugarcane bagasse is cellulose, which is difficult to decompose. In this study, natural cellulose-degrading bacterial strains were isolated and selected.
Methods: Bacteria were isolated from sugarcane bagasse in an environment containing carboxymethyl cellulose (CMC). The cellulase activity of microorganisms is reflected by the size of the halo zone on the medium containing CMC.
Results: Among the 8 isolates, three bacterial strains had strong resolving power (halo zones ≥ 20 mm). The strain with the maximum cellulolytic zone diameter was BH01, which exhibited the maximum zone of clearance around the colony with a diameter of 40 mm. The results of molecular identification showed that BH01 is a Bacillus amyloliquefaciens species with 100% similarity to the NICB. The optimal growth conditions for the BH01 strain were a pH of 7 and a temperature of 30°C.
Conclusion: Isolated bacterial strains with good cellulose degradation ability can be applied in practice.
INTRODUCTION
According to the Sugarcane Development Project to 2020, with a vision to 2030 of the Ministry of Agriculture and Rural Development, Vietnam's sugarcane production annually generates 7.7 million tons of bagasse, more than 1,137 million tons of molasses, and 1,149 million tons of sugarcane residue. Molasses is used to produce alcohol or for other microbial technologies. Sugarcane bagasse and mud residue contain nutrients that are used as a very good source of organic fertilizer for plants 1. However, the main component of sugarcane bagasse is cellulose. Cellulose can be hydrolyzed in alkaline or acidic environments. Physical and chemical methods for degrading cellulose are complex, expensive, and toxic to the environment. Treating organic waste containing cellulose with biotechnology and microorganisms with cellulose-degrading enzymes has many technical, economic and environmental advantages. Microorganisms that participate in the process of degrading cellulose under natural conditions are very diverse and include filamentous fungi, actinomycetes, bacteria and yeast 2.
Cellulase (EC 3.2.1.4; systematic name 4-β-D-glucan 4-glucanohydrolase) is a cellulose-degrading enzyme that can be found in many organisms, mainly fungi, bacteria, and protozoa. Cellulases are extremely important enzymes in industry and in the natural world, as they play a vital role in the global carbon cycle by breaking down insoluble cellulose into soluble sugars3. Bacillus species are recognized by many authors as strains with high cellulase production efficiency used in industry4, 5.
Extracellular cellulase is usually produced by various strains of microorganisms, including aerobic and anaerobic fungi and bacteria 6. Some microorganisms, such as spp. and spp., produce cellulase at high concentrations. species such as and have been widely used in cellulose production industries 4. spp. and spp. have also been found in many studies5. Some fungal species include sp. CR-316, sp., sp. and . Organisms such as protists, nematodes, mollusks, crustaceans and insects are also mentioned 7.
This study aimed to isolate and select bacterial strains capable of degrading cellulose from sugarcane bagasse to contribute to opening up a way to process sugarcane bagasse with many advantages in terms of technical, economic and environmental aspects.
MATERIALS AND METHODS
Materials
Reagents
Peptone, yeast extract, sodium chloride (Himedia), carboxymethyl cellulose (CMC), ammonium sulfate, potassium hydrophosphate, magnesium sulfate heptahydrate (Xilong Scientific), and TSI agar (Merck) were used. Gram stain kits were used: Lugol, violet, sagaphin, and Lugol (Hai Van Company).
Equipment
The materials used were vortexed (VELP Scientifica, Italy), refrigerated incubator shaker (New Brunswick Innova, Germany), and microscope (Leica, Germany).
Object
Sugarcane bagasse samples were collected from Bien Hoa, Tri An Sugar factory. The sugarcane stalks were cut quickly and stored in a 4°C refrigerator for further experiments.
Screening and isolation of bacteria
Based on previous studies 8, 9, bagasse samples were cut into pieces approximately 2-3 cm long. Sugarcane bagasse was layered into a 20 cm thick layer, 10% molasses was added to achieve 60% moisture, and the mixture was incubated for 5 days. During the incubation process, the samples were regularly checked, and molasses was added to ensure moisture. The samples to be isolated were diluted to the required concentration and subsequently cultured on basic nutrient media (peptone, 5 g/L; yeast extract, 2 g/L; NaCl, 1 g/L; agar, 15 gL; pH 6.5-7.0) to isolate the bacteria, (carboxymethyl cellulose). The use of Lugol as an indicator of cellulose degradation in agar media provides the basis for rapid and sensitive screening tests for cellulolytic bacteria10. Colonies showing discoloration of Lugol were considered positive cellulose-degrading bacterial colonies, and only those that showed discoloration were taken for further study. The obtained colonies were observed for colony morphology and Gram staining, and their motility and spore production were tested. The biochemical characteristics of the strains were tested via catalase and oxidase activity and via a sugar fermentation test using triple sugar iron (TSI) agar11.
Determination of cellulose degradation ability
The ability to dissolve cellulose was determined by the size of the halo zones on the carboxymethyl cellulose media (carboxymethyl cellulose, 10 g/L; (NH)SO, 1 g/L; KHPO, 1 g/L; MgSO.7HO, 1 g/L; NaCl, 1 g/L; pH 6.5). Bacteria were incubated for 5 days in a shaker incubator at 30°C and 100 rpm. Bacterial colonies capable of utilizing cellulose as the sole source of carbon were isolated on carboxymethyl cellulose agar media for 48 hours. Using 1% Lugor solution, the samples were spread on petri plates, after which the halo zone diameter was measured.
The resolving power (V) was calculated according to the formula:
V (mm) = D (mm) – d (mm)
where V is the resolving ability, D is the halo zone diameter, and d is the filter paper diameter.
The greater the resolving ability is, the greater the vitality of the extracellular space. The potency was classified according to the following criteria6:
V< 10 mm: Weak cellulose degradation ability (-),
15 mm >V ≥ 10 mm: Average cellulose degradation ability (+),
20 mm >V ≥ 15 mm: Good cellulose degradation ability (++);
V ≥ 20 mm: Strong cellulose degradation ability (+++)
Optimization of growth conditions for cellulose-degrading bacteria
To investigate the effects of temperature and pH on bacterial cell density, bacteria were inoculated in 250 mL of carboxymethyl cellulose media (pH 6.5). The culture mixture was incubated for 5 days at various temperatures (25°C, 30°C, 35°C, and 40°C). To study the effects of pH on bacterial cell density, the experiment was performed in carboxymethyl cellulose media at 30°C for 5 days. The pH was adjusted to values of 5, 6, 7, and 8 with 10% NaOH or 10% HCl solution. The bacterial cell density was determined using the critical dilution method.
Molecular identification of bacteria
Bacterial samples were sent for identification at Nam Khoa Company. The analysis method involved multiplication of the 16S rDNA gene sequence via PCR with the primer pairs 27F (5'AGAGTTTGATCCTGG CTCAG3') and 1492R (5΄- GGTTACCTTGTTACG ACTT-3'). BLAST software was used to compare the similarity of the sequence from the 16S rRNA region with the data in the GenBank of the National Center for Biotechnology Information (NCBI) to determine the species name of the bacterial strain.
Statistical analysis
All the experiments were performed in triplicate. Statgraphics Centurion XV software was used to statistically analyze the experimental data and evaluate the differences between the samples.
RESULTS AND DISCUSSION
Isolation of bacteria capable of degrading cellulose
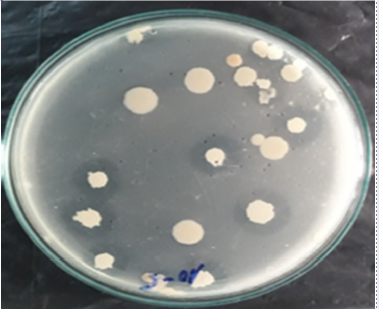
Sugarcane bagasse was cut into pieces approximately 2-3 cm long and was used as a raw material to isolate bacteria capable of degrading cellulose (Figure 1). After 5 days of incubation of the sugarcane bagasse in basic nutrient media, the culture solution was diluted in a decimal series from 10 to 10 with physiological saline. The cells were subsequently cultured on CMC isolation media. A bright area around the colonies indicates that the colonies are capable of degrading cellulose (Figure 2).

Raw material sugarcane bagasse

Colony morphology on CMC media
Initial results revealed that 8 bacterial strains were capable of degrading cellulose at concentrations of 10 and 10. After that, microbial strains capable of degrading cellulose were purified, kept and tested for biochemical properties. These strains were designated BH01 to BH08, respectively (
Characteristics of cellulose-degrading bacterial strains
|
Name |
Colony characteristics |
Morphological |
Spores |
Portability |
Catalase |
Oxidase |
TSI | |
|
Glu |
Lac, Suc | |||||||
|
BH01 |
Milky white, round shape, jagged edges |
gram-positive, long rod cells, 2.5-3 µm |
+ |
+ |
+ |
+ |
+ |
+ |
|
BH02 |
Pale yellow, slimy, flat, uneven edges |
gram-positive, short rod cells, 1 µm |
- |
- |
+ |
+ |
+ |
+ |
|
BH03 |
Milky white, wrinkled, uneven |
gram-positive, short rod cells, 1 µm |
+ |
+ |
+ |
+ |
+ |
- |
|
BH04 |
Milky white, irregularly round, slimy |
gram-negative, short rod cells, about less than 1 µm |
+ |
+ |
+ |
+ |
+ |
- |
|
BH05 |
Milky white, rough and wrinkled face |
gram-negative, short rod cells, 1-1.5 µm. |
- |
+ |
+ |
+ |
+ |
+ |
|
BH06 |
Milky white, evenly round, concentric rings, slimy |
gram-positive, short rod cells, 1.5-2 µm |
+ |
+ |
+ |
+ |
+ |
+ |
|
BH07 |
Light yellow, slightly slimy, evenly rounded edges |
gram-negative, short rod cells, 1.5-2 µm |
- |
- |
+ |
+ |
- |
- |
|
BH08 |
Milky white, irregularly round, slimy |
gram-positive, rod cells, 2-3 µm |
+ |
+ |
+ |
+ |
+ |
+ |
The cells were observed under a microscope at 1000x magnification. Gram staining revealed that the isolated bacterial strains were both gram-negative and gram-positive. In terms of morphology, the cells had a straight rod shape, single cells, and pairs; the cell diameter ranged from 0.4 -1.2 µm, and the length ranged from 1 - 3 µm. Of the 8 isolated bacterial strains, 5 strains were capable of producing spores, and 6 strains were portable. All the isolated strains had catalase and oxidase activities. TSI agar containing three types of sugars (glucose, lactose and sucrose) was used. This test was performed to determine the possible use of different carbohydrate resources in the basal growth media. Test results showed that among the 8 isolated bacterial strains, 7 were able to use glucose, and 5 were able to use lactose and sucrose.
Determination of cellulose degradation ability
The above 8 bacterial strains were cultured from the test tube in carboxymethyl cellulose media with shaking for 5 days. An autoclaved round filter paper (1.5 cm in diameter) was placed on a CMC agar plate. A pipette was used to aspirate 40 µL (enough volume to absorb into the filter paper) of the bacterial solution with a cell density of 10 CFU/mL after shaking for enrichment onto filter paper; then, the bacterial solution was allowed to breach into the environment. After 48 hours, 1% Lugor solution was used to spread the cells on the Petri dish, after which the halo zone diameter was measured. The results are shown in
Results of testing the ability to degrade cellulose by measuring the resolution cycle of bacterial strains isolated on CMC media.
|
Name |
The halo zones diameter |
Resolution |
|
BH01 |
40.00 |
+++ |
|
BH02 |
20.33 |
+++ |
|
BH03 |
19.33 |
++ |
|
BH04 |
11.33 |
+ |
|
BH05 |
12.67 |
+ |
|
BH06 |
10.67 |
+ |
|
BH07 |
20.00 |
+++ |
|
BH08 |
14.00 |
+ |

(A) Cell shape of the bacterial strain BH01 under a 100× objective lens; (B) Halo zones of strain BH01 on CMC medium
The results of the experiment are also relatively consistent with those of a number of other studies. Author NTT Thuy (2018) collected soil samples from landfills, sawdust factories, and cultivated lands at 12 locations in the Phu Vang, Huong Tra, Phong Dien, and Quang Dien districts and the city. Hue, Thua Thien Hue Province. The results revealed a microbial strain with strong cellulose degradation ability and a resolution circle diameter larger than 20 mm10.
NNT Ngan . (2014) isolated strains of bacteria, actinomycetes, and molds capable of degrading cellulose from cassava peel waste at the Thua Thien Hue cassava starch factory. The results of the study showed that there are 2 mold strains with a diameter of 27-28.5 mm in cellulose degradation on CMC media, two actinomycete strains with a resolution diameter of 21-24 mm, and 2 bacterial strains with a cellulose degradation diameter of 23-24.5 mm12.
Do Nang Vinh (2020) isolated sugarcane soil samples and compost from straw in Ha Tinh. The cellulose degrading activity of the isolated strains ranged from 8 to 30 mm, and actinomycetes are the group with the highest degrading activity13.
Optimization of growth conditions for cellulose-degrading bacteria
To investigate the effects of temperature and pH on bacterial cell density, 5 ml of the bacterial growth solution BH01 at a density of 10 was inoculated into 250 ml of carboxymethyl cellulose media. To investigate the effect of temperature on bacterial cell density, different incubation temperatures of 25°C, 35°C, 35°C and 40°C were used. After 5 days of culture, the number of bacterial cells was determined using the critical dilution method.
According to the results shown in
Effect of temperature on the bacterial cell density
|
Temperature (0C) |
Cell density (CFU/mL) |
|
25 |
(2,5x108)a |
|
30 |
(8,2x109)b |
|
35 |
(7,5x109)b |
|
40 |
(6,2x109)c |
To investigate the effect of environmental pH, the experiment was performed in carboxymethyl cellulose media at 350°C for 5 days, similar to what was previously described. The pH was adjusted to 5, 6, 7 and 8 with 1% NaOH or 1% HCl solution.
Effect of pH on bacterial cell density
|
pH |
Cell density (CFU/mL) |
|
5 |
(1,5x106)a |
|
6 |
(6,2x107)b |
|
7 |
(8,7x109)c |
|
8 |
(7,5x109)c |
Molecular identification of bacteria
Identification of the bacterial strain BH01 by 16S RNA gene sequencing Bacterial samples were sent for identification at Nam Khoa Company. The analysis method involved multiplication of the 16S rDNA gene sequence via PCR with the primer pairs 27F (5'AGAGTTTGATCCTGG CTCAG3') and 1492R (5΄- GGTTACCTTGTTACG ACTT-3'). BLAST software was used to compare the similarity of the sequence from the 16S rRNA region with the data in the GenBank of the National Center for Biotechnology Information (NCBI) to determine the species name of the bacterial strain. This sequence was subsequently checked via a gene bank through BLAST, and the results showed that the bacterium BH01 is a member of the species with 100% similarity to the NICB (number CP031424.1). This result is also consistent with previously analyzed bacterial cell characteristics. Based on the NCBI data analysis and the submission of the 16S rRNA gene sequence, a phylogenetic tree was constructed for Bacillus amyloliquefaciens BH01 (Figure 4).

Phylogenetic tree of
This result is consistent with the findings of many other studies. is a strain capable of producing strong cellulase enzymes. Sun, L et al. (2017) isolated the strain S1 from the cecum of geese. The enzyme activity of the strains was determined by the reducing sugar method, and appropriate culture conditions for producing cellulase showed that the bacteria had a high cellulase activity of 1.25 U/mL14.
In Vietnam, NN An et al. (2021) cultured D19 in an environment with a pH 5 supplemented with the following media ingredients: 0.5% straw, 1% soybean meal, 1% NaCl, and 0.5% yeast extract after 48 hours of shaking at 150 rpm and 37°C. The enzyme activity reached its highest value (879.47 UI/ml) 15.
CONCLUSIONS
Eight strains of bacteria capable of degrading cellulose were isolated from bagasse samples collected from Bien Hoa, Tri An Sugar Factory. The results of testing cellulase activity through the halo zones on CMC agar medium showed that 3/8 of the bacterial strains had strong dissolution ability (V ≥ 20 mm). Strain BH01 has the highest resolution ring diameter of 40 mm and was selected for species identification. The results showed that the bacterium BH01 belongs to the species and has 100% similarity to NICB. Strain BH01 has optimal growth conditions at pH 7-8 and a temperature range of 300C-35C.
LIST OF ABBREVIATIONS
CMC: carboxymethyl cellulose
NCBI: National Center for Biotechnology Information
TSI: triple sugar iron
COMPETING INTERESTS
The authors declare that they have no competing interests.
ACKNOWLEDGEMENTS
This study was conducted at the laboratories of the Department of Biotransformation Technology, Institute of Tropical Biology, Ho Chi Minh City, Vietnam.
Identification of microorganisms using molecular biology methods at Nam Khoa Company
AUTHOR’S CONTRIBUTIONS
All the authors contributed to designing and conducting the experiments, analyzing and interpreting the data and drafting and revising the manuscript.

