Biofilm biosynthesis of Priestia megaterium M1VB5 strain Muc Son Thanh Hoa paper mill
- Institute of Biotechnology, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet, Cau Giay, Hanoi, Viet Nam
- Faculty of Environmental Sciences, University of Science, Vietnam National University, Hanoi, 334 Nguyen Trai, Thanh Xuan, Hanoi, Viet Nam
- Research institude of pulp and paper industry, No 59 Vu Trong Phung, Thanh Xuan, Hanoi, Viet Nam
Abstract
Introduction: Strain M1VB5, isolated from slime in paper machine pipes, is studied for its role in slime formation. This slime, comprised of microorganisms, binds and adheres to pipes, potentially leading to clogs, process inefficiencies, and machinery contamination.
Methods: We investigated the biological characteristics and gene sequencing of M1VB5, focusing on its biofilm biosynthesis capabilities under varying sucrose concentrations, temperatures, pH levels, and time periods.
Results: M1VB5 exhibited adaptability across a broad range of conditions, thriving at temperatures between 15-55°C and pH levels from 5 to 9, with up to 7% salinity tolerance. Capable of utilizing diverse carbon sources, M1VB5 efficiently biosynthesized extracellular enzymes, including CMCase, cellulase, xylanase, amylase, protease, and chitinase. Identified as Priestia megaterium (Gram-positive), it demonstrated significant biofilm formation, producing 36.4 g/L EPS and forming a 53.73 g/100g biofilm on PVC at a sucrose concentration of 100 g/L. Optimal biofilm production was observed at pH 8, with 28.40 g/L EPS and a biofilm mass of 59.61 g/100g PVC, and at 37°C, where EPS and biofilm weights reached 23.60 g/L and 63.78 g/100g PVC, respectively. The strain maintained steady biofilm production from day 4 to 14.
Conclusion: M1VB5's versatile carbon source usage, wide temperature and pH range tolerance, and diverse enzyme production facilitate its growth and development. Its significant biofilm and EPS production capabilities contribute to its dominance in slime formations, offering insights for managing slime in industrial paper mill environments.
INTRODUCTION
Slime, commonly referred to in paper mills, originates from microbial deposits that include bacteria, their exopolysaccharides (EPS), fibers, and various additives involved in paper production 1. The process begins with the accumulation of biofilm and EPS-producing bacteria on paper mill equipment, which adheres and multiplies on surfaces 2, 3. Subsequent layers of microorganisms and production materials attach to the initial biofilm, enlarging the slime mass. EPS, vital for the structural integrity of biofilms, facilitates microbial interactions and adherence, leading to the slime's mucous and viscous nature4.
This occurrence is a natural consequence of microbial growth in environments with suitable nutrition, humidity, and temperature. Closed paper mill systems that recycle fibers create ideal conditions for such microbial proliferation, given the optimal nutrient availability, temperature, and pH3. The microbial spectrum in slime encompasses bacteria from various genera, including Aerobacter, Micrococcus, Serratia, Ascophyta, Clonostachys, Chlamydobacteria (bacteria with a membrane), Bacillus, Aspergillus, Cephalosporium, Penicillium, Fusarium, Rhodotorula, Monilia, yeast, and Pseudomonas5, 6.
While these biofilm-producing microbial strains are problematic in paper mill operations, they offer benefits in other industries, such as the livestock, food, and oil spill remediation sectors. Nonetheless, studies focusing on microbial biofilm synthesis in aquatic environments are limited. This paper introduces the biological characteristics and biofilm production of the M1VB5 strain, isolated from the slime of the Muc Son - Thanh Hoa paper mill, underscoring the importance of further research in this field.
MATERIALS AND METHODS
Bacterial strain
TheM1VB5 strain was isolated from slime sample on paper production machine collected at Muc Son Paper mill - Thanh Hoa.
Media
(g/L): Sucrose 20; yeast extract powder 0.5; (NH)SO 0.6; KHPO 3.18; KHPO 5.2; MgSO.7HO 0.3; CaCl2HO 0.05; ZnSO.7HO 0.0002; CuSO.5HO 0.0002; MnSO.HO 0.0002; CoCl.6HO 0.0002; FeSO.7HO 0.0006; pH 7.0, agar 20.0 7.
(g/L): Mannitol 10.0; KHPO 0.5; MgSO.7HO 0.2; NaCl 0.1; yeast extract powder 0.5; pH 7.0.
Determination of some growth characteristics
The M1VB5 strain was cultured at temperatures from 0-55C, pH from 3 to 10, salt concentration from 0 -10% and different sources. From there, suitable growing conditions can be determined 8.
Determination of extracellular enzyme production
The M1VB5 strain was dotted on Petri dishes containing Gause I media with the addition of specific substrates (1%): starch to determine amylase activity; casein to determine protease activity; chitin to determine chitinase activity; CMC and cellulose to determine cellulase activity; xylan to determine xylanase activity. After 5 days, determine the substrate decomposition ring with reagents, lugol with plates with the addition of starch, CMC (Carboxymethyl Cellulose) and cellulose; protease with 10% tricloacetic reagent; chitinase with red Congo reagent 0.5%. The ability to biosynthesize enzymes is determined by the D-d value (mm), where D is the substrate decomposition ring diameter, and d is the colony diameter 8.
Investigation of slime-forming ability of the M1VB5 strain
PVC plates measuring 20 x 20 mm were suspended in a Sucrose-ammonium medium. After an appropriate amount of time, PVC plates are removed from the medium for analysis. The collected plates were weighed to determine the newly formed biofilm content on PVC plates9.
Determination of optimal culture conditions for biofilm biosynthesis
- : The M1VB5 strain was fermented on Sucrose-ammonium medium supplemented with varrying amounts of sucrose 50; 75; 100; 120; 150; 170; 200; 250 (g/L) (other factors remain unchanged). After 7 days of fermentation, the weight of biofilm and the content of EPS were determined.
- : The M1VB5 strain was fermented on an optimal medium for biofilm biosynthesis at different pHs: 4, 5, 6, 7, 8, 9, 10 and 11(other factors remain unchanged). After 7 days of fermentation, the weight of biofilm and the content of EPS were determined.
: The M1VB5 strain was fermented on an appropriate medium and pH at varrying temperatures: 20, 30; 37; 45 and 50C(other factors remain unchanged). After 7 days of fermentation, the weight of biofilm and the content of EPS were determined.
Determination of EPS content: The M1VB5 strain was fermented in sucrose-ammonium medium, at 30oC, 120 rpm for 2 days. The fermentation fluid was centrifuged at 12000 rpm for 15 min to remove biomass. Add cold ethanol (leave overnight at -20°C), precipitate EPS in the ratio of ethanol:fermentation fluid = 3:1 (v/v), stir slightly and keep at -20°C for 1 hour. Centrifuged at 8000 rpm for 15 minutes at 4°C. Remove floating liquid, scale and dry at 60 °C for 48 hours until constant volume. Determine the volume of the scale and calculate the result 10.
Quantification of biofilm
Quantification of biofilm production was performed on 96-well disks.. The steps are summarized as follows: (i) culture bacteria in liquid medium using 96-well plate at 37C within 24 hours; (ii) rinse off any non-adhered bacteria with PBS 1x; (iii) stain the biofilm with a 1% crystalline violet solution; (iv) wash to remove excess crystalline violet solution; (v) fix the biofilm with methanol for 20 minutes at room temperature; (vi) determine the optical density of each well at a wavelength of 630 nm to evaluate the degree of biofilm formation 11.
DNA extraction and gene sequencing analysis 16S rRNA
Total DNA of M1VB5 strain was extracted using the method of Sambrook and Russell (2001) 12. The 16S rRNA gene was amplified by Polymerase Chain Reaction (PCR) using a primer pair 27F (5'-TAACACATGCAAGTCGAACG-3') and 1492R (5'-GGTTACCTTGTTACGACTT-3')13. PCR reaction occurs in 35 cycles, each cycle is as follows: 94C for 5 minutes, 94C for 90s, 51C for 30s, 72C for 90s, 72C for 10 minutes. PCR products were visualized on 1% agarose gel and purified using the PureLinkTM – DNA Purification (Invitrogen) kit. The 16S rRNA sequence was analyzed and sequenced using the DNA Sequencing/Capillary Electrophoresis method of Apical Scientific Laboratory. The 16S rRNA gene sequence were compared with other sequences on GenBank bank using BLAST (www.ncbi.nlm.nih.gov).
RESULT
Biological characteristics of M1VB5 bacteria
Biological characteristics of M1VB5 strain
|
Growth characteristics |
Substrate decomposition ring, D (mm) | ||
|
Salinity tolerance |
0 – 7 % |
CMC |
27.0±0.67 |
|
Growth temperature |
15 – 55oC |
Cellulose |
8.0±0.33 |
|
Growth pH |
5 - 9 |
Xylan |
25.0±1.00 |
|
Ability to use carbon sources |
D-glucose, L-arabinose, D-xylose, D- fructose, lactose, D-rafinose, sucrose, D- xylose |
Soluble starch |
14.0±0.67 |
|
Hydrolyzed casein |
28.0±1.33 | ||
|
Chitin |
25.0±1.33 | ||
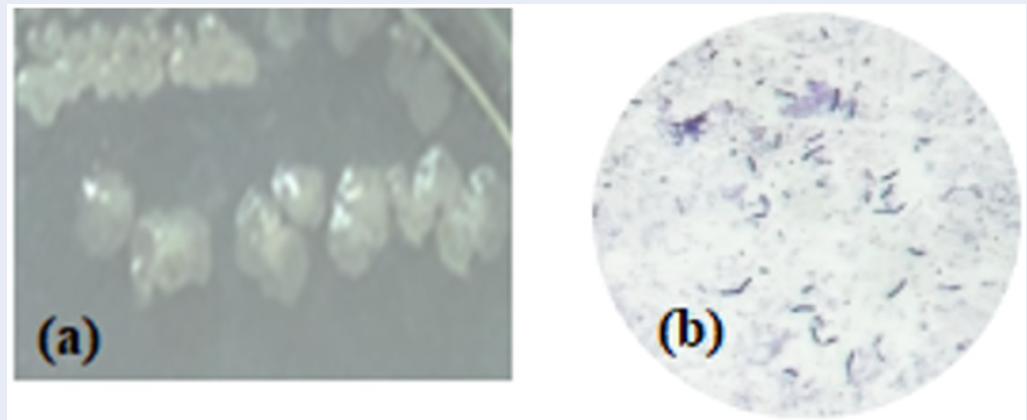
The M1VB5 strain formed colonies on the isolated medium with characteristics including mucus texture, a glossy apperance, round and spherical convex shape with milky coloration, an even colony margin with a diameter of 1-2mm, central lobed dissection, and concentric wrinkles (Figure 1a). In addition, the bacteria has a long rod-shaped and Gram (+) stained (Figure 1b). The M1VB5 strain was able to grow at temperature range of 15 – 55C and across pH levels of 5-9, with optimal conditions observed at 25 – 37C and pH 6-8. The ability of M1VB5 to decompose a variety of substrates such as CMC, cellulose crystals, xylene, soluble starch, casein, chitin was also demonstrated (Figure 2). Moreover, M1VB5 bacteria was capable of using a variety of carbon sources: D-glucose, L-arabinose, D-xylose, D-fructose, lactose, D-rafinose, and sucrose (

Cell morphology on culture plates (a) and Gram stain morphology (b)

Ability to decompose some substrates of M1VB5 strain (a) CMC; (b) Cellulose; (c) Xylane; (d) Soluble starch; (e) Casein; (f) Chitin

Effect of pH on M1VB5's slime-forming ability after 7 days

Biofilm biosynthesis over time of M1VB5 strain

Phylogenetic tree of strain M1VB5
Effect of sucrose and culture conditions (temperature and pH) on biofilm biosynthesis of M1VB5 strain
The M1VB5 strain produced biofilm across various sucrose concentrations, with optimal production observed at sucrose concentrations of 100 g/L and 200 g/L. At a sucrose concentration of 100 g/L, strain M1VB5 had a biosynthesis capacity of 36.4 g/L EPS and biofilm of 53.73 g/100g PVC. Similarly, at a sucrose concentration of 200 g/L, the EPS content of 39.2 g/L and biofilm of 53.67 g/100g PVC were obtained from M1VB5.
Effects of sucrose, temperature and pH on EPS and biofilm biosynthesis of M1VB5
|
Effect of sucrose content on EPS and biofilm production | ||||||
|
Sucrose content (g/L) |
50 |
100 |
150 |
200 |
250 | |
|
EPS content (g/L) |
10.8±0.18 |
36.40±0.21 |
37.2±0.19 |
39.2±0.24 |
36.0±0.34 | |
|
Biofilm weight (g/100g PVC) |
29.08±0.25 |
53.73±0.34 |
39.05±0.12 |
53.67±0.39 |
39.24±0.24 | |
|
Effect of pH on EPS and biofilm production | ||||||
|
pH |
5 |
6 |
7 |
8 |
9 |
10 |
|
EPS content (g/L) |
14.4±0.17 |
27.6±0.19 |
22.4±0.15 |
28.40±0.21 |
6.4±0.14 |
8.0±0.16 |
|
Biofilm weight (g/100g PVC) |
20.01±0.26 |
45.10±0.22 |
56.94±0.13 |
59.61±0.23 |
17.80±0.17 |
26.28±0.22 |
|
Effect of temperature on EPS and biofilm production | ||||||
|
Temperature (oC) |
20 |
30 |
37 |
45 |
55 | |
|
EPS content (g/L) |
17.20±0.15 |
18.00±0.21 |
23.60±0.24 |
21.26±0.26 |
16.00±0.21 | |
|
Biofilm weight (g/100g PVC) |
7.60±0.18 |
46.54±0.18 |
63.78±0.22 |
38.52±0.29 |
15.19±0.20 | |
The M1VB5 strain displayed robust growth within the pH range of 6 to 8, at different pH the culture colour also varies, from light yellow to dark yellow and pink (Figure 3). Furthermore, within this pH range, the M1VB5 strain exhibited maximum biofilm production, reaching an EPS content of 28.40 g/L at pH 8 and a biofilm weight of 59.61g/100g PVC.
The M1VB5 strain demonstrated significant biofilm biosynthesis at the optimal temperature at 37°C, reaching an EPS content of 23.60 g/L and a biofilm content of 63.78 g/100g PVC. Since the typical ambient temperature in paper mills and numerous food manufacturing plants is around 37°C, this bacterium poses a risk of biofilm formation in such facilities. However, environmentally, this bacterial strain is also considered safe and capable of biosynthesizing numerous active ingredients, making it applicable in various biotechnological fields 14.
Biofilm biosynthesis over time of M1VB5 strain
Experiments conducted in 96 wells demonstrated that the M1VB5 strain exhibited a high capacity for biofilm production, maintaining stability after 4 days and up to 14 days. The biofilm content remained relatively stable, ranging from 0.73 to 0.83 at OD 630nm (Figure 4). This finding is crucial in guiding biofilm treatment in paper production line at paper mills.
Identification of M1VB5 strain by 16S-rRNA gene analysis
The 16S-rRNA gene sequence of the M1VB5 strain with a size of about 1264 base pairs was compared with strains published on the Genbank database. The M1VB5 16S rRNA sequence had high similarities (99.9%) compared to the corresponding genes of , DSM 32, IAM 13418, NBRC 15308. These results suggest that M1VB5 is identified as M1VB5.
DISCUSSION
Our study of the . M1VB5 strain has led to several significant findings regarding this bacterium. While there are differences between our research strain and those described by previous authors, notable similarities also exist. As detailed by De Vos et al. (2009), . is characterized as a rod-shaped, Gram-positive, predominantly aerobic, spore-forming bacterium found in a variety of habitats15. It is notably large for a bacterium, with cell lengths of up to 4 μm and diameters of 1.5 μm. Typically, the cells form pairs or chains, linked by polysaccharides on their cell walls15, 16. can grow in temperatures ranging from 15 to 45°C, with an optimal temperature of 30°C. However, strains isolated from Antarctica's geothermal lakes have demonstrated the ability to grow at temperatures as high as 63°C15. The M1VB5 strain of can grow in a broader temperature range, from 15 to 55°C, showcasing its heat resistance compared to other strains. This strain is utilized as a probiotic in feeds for fish and shrimp, thanks to its production of cellulase and amylase enzymes, measured at 3.97 U/mL and 1.83 U/mL, respectively17.
Verhoef et al. (2005) have suggested that D-glucose or D-sucrose are the primary substrates for exopolysaccharide (EPS) production in the Bacillus genus18. Our findings indicate that the M1VB5 strain efficiently utilizes D-glucose and D-sucrose. In the context of paper production machinery, where conditions favor the growth of M1VB5, this can lead to rapid slime formation. This poses a challenge in the food processing industry, where sucrose is extensively used, as it serves as a nutrient source that facilitates biofilm formation, adversely affecting food quality. According to Chaudhary . (1997), most biofilm-forming microorganisms thrive at neutral pH and 37°C, but their growth is inhibited at temperatures above 50°C, which helps limit biofilm formation19.
The accumulation of biofilm over time is a critical aspect, with its mass increasing progressively until it detaches and relocates, beginning the accumulation process anew. This cycle of slime formation and detachment is a continuous challenge, particularly in papermaking, where untreated slime can significantly impact product quality and the production process20. The treatment of slime, often requiring the cessation of machine operations and the use of hot solutions, not only poses environmental concerns but also reduces productivity due to downtime. Identifying the optimal timing for the treatment of significant slime accumulations remains a pressing challenge, crucial for maintaining product quality and environmental sustainability.
CONCLUSION
The M1VB5 strain exhibited growth within a temperature range of 15 – 55°C, with optimal growth occurring at 25 – 37°C, and a pH range from 5-9. This strain has the capability to decompose various substrates, including CMC, cellulose, xylan, soluble starch, casein, and chitin. With a high genetic similarity (99.9%) to the species, M1VB5 has been identified as M1VB5. The M1VB5 strain demonstrated significant biofilm biosynthesis, especially with sucrose concentrations of 100-200 g/L. Notably, it produced substantial biofilm at pH 8, with an EPS content of 28.40 g/L and a biofilm weight of 59.61 g/100g PVC. Moreover, at a temperature of 37°C, the EPS content reached 23.60 g/L, and the biofilm weight reached 63.78 g/100g PVC. This strain is also capable of robustly synthesizing biofilms and maintaining relatively constant production from day 4 up to day 14. With the results achieved, we have gained an overview of slime production, from which we can explore methods for its treatment. With the detection of the M1VB5 strain, it proves useful not only for the paper industry but also for other industries, such as the food industry and oil propagation treatment technology.
Abbreviations
None.
Author's contributions
All authors contribute to the conception and design of the study. PTHT: Conceptualization, methodology, formal analysis, evaluation and correction. DTN, TTH, LTT, NTHL: ideation, formal analysis, investigation, writing – original manuscript. NVH, CVS: experimental sampling, factory survey. TBKN: review and edit. All authors have read and approved the final manuscript.
Competitive interests
We ensure there is no competition of interest among authors.
Acknowledgments
This research received funding from the project: "Research on creating biodispersion preparations applied to microbial slime treatment in industrial packaging paper production lines". Code: 016.2022.ĐT.BO/H DKHCN under the project of the Ministry of Industry and Trade.

