Molecular detection of Escherichia coli O157:H7 isolates from stool samples in Khartoum State, Sudan
- Faculty of Medical Laboratory Sciences, National University-Sudan
- Department of Microbiology, Ibn Sina University, Sudan
- Department of Microbiology, Omdurman Ahlia University, Sudan
- Ahfad University for Women
Abstract
Background: E. coli O157:H7 is a causative agent of food-borne illness. This study aimed to detect the presence of E. coli O157:H7 in stool samples from Khartoum by using a molecular technique. This was a laboratory-based study. A total of 100 stool samples were collected from different types of hospitals in Khartoum State; 54 (54%) male and 46 (46%) female samples were cultured on Mac- Conkey agar media at 37◦C for 24 hours overnight, and colony morphology, Gram's staining technique and different biochemical tests were subsequently performed for identification.
Results: The results revealed that E. coli was present in 70% of the samples analyzed. DNA was extracted via the boiling method, and primers for E. coli O157:H7 were used to detect the presence of genes via the polymerase chain reaction (PCR) technique. Molecular detection via PCR was performed on 70 isolates of E. coli to detect the target gene O157:H7, and the results revealed that a band appeared in 1 (1.4%) E. coli O157:H7 sample and that no band was detected in 69 (98.6%) samples.
Conclusions: The study concluded that the isolation of E. coli O157:H7 from patients should direct the attention of physicians to the possibility of complications among young and elderly patients.
Background
are gram-negative bacilli, catalase test positive, oxidase test negative, facultative anaerobic, some strains capsulated, flagellated, citrate test negative and indole test positive. It is normally found in the gastrointestinal tract of humans and animals. Some of these diseases cause diarrhea and a range of extraintestinal diseases 1.
has a broad spectrum of lifestyles and phenotypes and is a well-suited model organism for studying bacterial evolution and adaptation to different growth conditions 2. A harmless commensal needs only to acquire a combination of mobile genetic elements to become a high pathogen capable of causing a range of diseases 3.
Enteric is both a normal flora and a pathogen that causes morbidity and mortality worldwide. The strains that cause gastroenteritis are traditionally divided into 6 pathotypes on the basis of their virulence properties, mechanisms of pathogenicity, clinical syndromes and O:H serogroups: enteropathogenic (EPEC), enterotoxigenic (ETEC), enteroinvasive (EIEC), and enteroaggregative (EAEC) enterohemorrhagic (EHEC) 4.
Infectious bacteria can be spread from humans and animals through the consumption of uncooked meat, the consumption of contaminated fruits and vegetables, the consumption of unpasteurized milk, and contact with infected animals; anyone has the potential to develop infection if they are exposed to the bacteria, and symptoms may begin 1 to 10 days after exposure5.
Most infections are opportunistic infections. Special strains of can cause diarrhea, one of which is verotoxigenic . 6.
The entero-hemorrhagicrhaic (EHEC) serotype O157:H7 is the most commonly identified member of the Shiga toxin-producing (STEC) family; it is the most notorious emerging pathogen and is considered prototypical for the current paradigm of foodborne disease in the United States7. It is a human pathogen responsible for bloody diarrhea, causing sporadic and epidemic outbreaks and severe life-threatening hemolytic uremic syndrome (HUS) worldwide8. Previously isolated as a foodborne pathogen in 1982 in the United States (U.S.), a rare strain of was investigated and isolated from stool and food samples. Serotyping confirmed the presence of the somatic O157:flagellar H7 9. O157:H7 is a cause of intestinal disorders, ranging from mild infection to severe bloody diarrhea. Most isolates have been shown to produce Shiga toxins (Stx1, Stx2), and most clinical laboratories now routinely screen stools for O157:H7 from patients with bloody and nonbloody diarrhea10. O157:H7 infects the alimentary tract, induces abdominal cramps with hemorrhagic diarrhea and causes a wide range of clinical illnesses, including nonbloody diarrhea, hemorrhagic colitis, hemolytic uremic syndrome and death. The infection is initiated by the ingestion of a small inoculum of 10–100 CFUs 11.
O157:H7 is a causative agent of foodborne illness through the consumption of contaminated milk and uncooked beef and may be present after swimming in or drinking water contaminated with O157:H7.
Infection with these bacteria causes hemorrhagic diarrhea and kidney failure, leading to hemolytic uremic syndrome (HUS) according to the Central Disease Control (CDC) (3–5%), and individuals who develop hemolytic uremic syndrome (HUS) may die from this complication.
This study aimed to detect O157:H7 in stool samples from Khartoum State, Sudan
Methods
Study Design:
This laboratory-based study was performed in different hospitals in Khartoum state (Al-Amal National Hospital, Jabal Awliya Hospital, Bahri Aleaskarii Hospital). The sample size included all samples from hospitals during the study period. The inclusion criterion included all patients with gastrointestinal distress, whereas the exclusion criterion did not include patients without gastrointestinal distress.
Sample collection:
Stool samples were collected from a total of 100 diarrheic patients in wide-mouth containers from different hospitals.
Direct wet preparation:
One drop of normal saline was placed on a clean slide.
- A small portion of each stool sample was collected via a wound stick and mixed well with a drop of normal saline.
The glass was covered and then examined under a microscope with 10x and 40x objectives12.
Culture technique:
Diarrheal samples were cultured on MacConkey agar and incubated for 24 hours at aerobically 13.
Identification of bacteria:
The isolated organisms were identified via standard laboratory procedures via colony morphology, the Gram's staining technique and different biochemical tests.
Colonial morphology:
The colonies on MacConkey agar were lactose fermented, medium-sized, round, smooth, slightly convex and pink in color. Yellow colonies (nonlactose ferment) were excluded 14. The Gram staining technique was performed according to a known protocol in the literature15.
Biochemical tests:
These tests include the use of indole in tryptone water16, citrate utilization tests17, urease tests, Kligler iron agar (KIA) 18, and motility tests (nonbiochemical tests) 19, 20.
Molecular technique
Genomic DNA was extracted from whole cells via the heat-shock method in this study. Twenty-four-hour colonies were grown on nutrient media, and the culture media were harvested in 2 ml of sterile distilled water with a sterile loop. One millimeter aliquot of the cell suspension was transferred to a 1.5 ml microcentrifuge tube, and the cell mixture was boiled in a water bath for 10 minutes, followed by sudden freezing for 10 minutes. This step was repeated three times. The cell debris was pelleted by centrifugation at high speed (1300 rpm at 4°C for 10 minutes), and the supernatant was transferred to a new 1.5 ml tube and then used immediately as a template for PCR amplification or kept at 4°C for up to one month (Santa Cruz Biotechnology Dallas Company, TX, USA) 21.
Polymerase chain reaction (PCR):
The following steps were adopted to carry out the process of DNA amplification. Five microliters of the extracted DNA was added to the Maxime PCR premix kit (i-Taq) mixture, and 1 μL of each forward or reverse primer mixture (1 bp) plus 13 μL of distilled water was added. Thermal cycling for 35 cycles was performed at 94°C for 1 min, 60°C for 1 min and 72°C for 5 min. The final extension step was performed for 5 min at 72°C (Santa Cruz Biotechnology Dallas Company, TX, USA).
DNA inhibitors were removed as described previously, and the manufacturer’s guidelines were precisely followed by the use of master mix for PCR primers (Santa Cruz Biotechnology Dallas Company, TX, USA)22.
Primers:
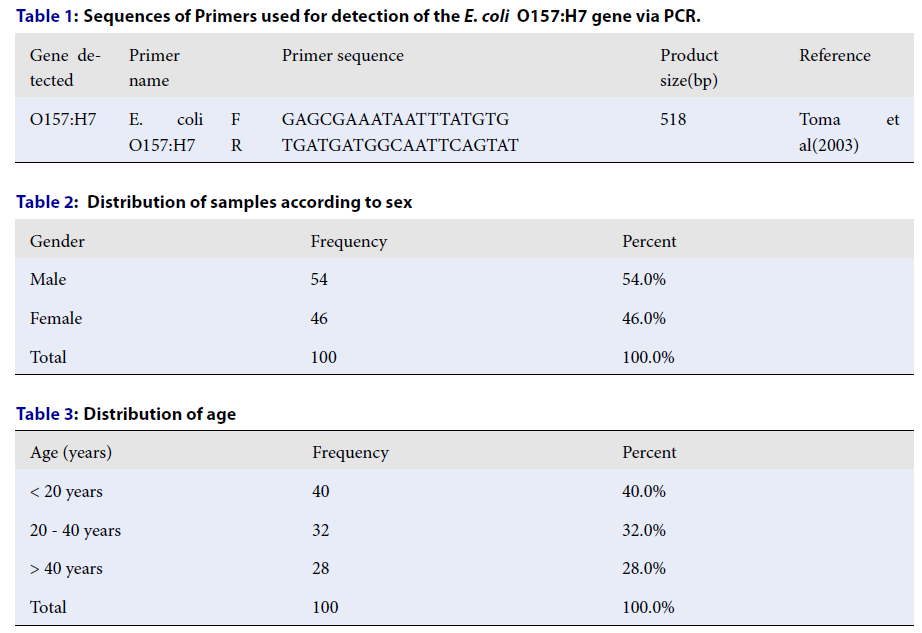
Sequences of Primers used for detection of the
|
Gene detected |
Primer name |
Primer sequence |
Product size(bp) |
Reference | |
|
O157:H7 |
E. coli O157:H7 |
F R |
GAGCGAAATAATTTATGTG TGATGATGGCAATTCAGTAT |
518 |
Toma et al(2003) |
A 10% agarose gel 23 was prepared, and gel electrophoresis was performed via a documentation system (Bio-Rad, USA) with a run time of 6 hours24, 25.
Data analysis:
The data were analyzed via the Statistical Package for Social Sciences (SPSS) version 20.
Results
The present study addresses a significant public health concern: the detection of O157, a notorious food-borne pathogen, in stool samples from Khartoum State. The use of molecular techniques, specifically polymerase chain reaction (PCR), aims to assess the prevalence of this pathogen among patients and draw attention to its potential health implications. Given the relevance of food safety and disease prevention in the context of increasing global health threats, this study provides important insights.
A total of 100 diarrheal samples were collected from people of different ages in Khartoum state, ranging from 9 months to 70 years old, who can be male or female during the study period (
All the samples were cultured on MacConkey agar, and colony morphology, Gram's staining technique and different biochemical tests were performed. The results revealed that accounted for 70 (70%) samples, whereas accounted for 30 (30%) samples (
The distribution between the gander and test results revealed that 38 (38%) males and 32 (32%) females were positive for (
Molecular detection via PCR was performed on 70 isolates of for detection of the target gene O157:H7, and the results revealed that a band appeared in 1 (1.4%) O157:H7 sample and that no band was detected in 69 (98.6%) samples, which are considered other than O157:H7 (
The associations between sociodemographic factors (gender, age) and test results revealed that 1 (1.4%) male was positive for O157:H7 via the PCR technique, and the other 37 (52.9%) females and 45 males (45.7%) were negative for O157:H7 (P value = 0.488) (
Distribution of samples according to sex
|
Gender |
Frequency |
Percent |
|
Male |
54 |
54.0% |
|
Female |
46 |
46.0% |
|
Total |
100 |
100.0% |
Distribution of age
|
Age (years) |
Frequency |
Percent |
|
< 20 years |
40 |
40.0% |
|
20 - 40 years |
32 |
32.0% |
|
> 40 years |
28 |
28.0% |
|
Total |
100 |
100.0% |
Distribution of
|
Frequency |
Percent | ||
|
Culture for |
Positive |
70 |
70.0% |
|
Negative |
30 |
30.0% | |
|
Total |
100 |
100.0% | |
Association between gender and test results
|
Gender |
Total |
P. value | |||
|
Male |
Female | ||||
|
Culture for E. coli |
Positive |
38 (38.0%) |
32 (32.0%) |
70 (70.0%) |
0.930 |
|
Negative |
16 (16.0%) |
14 (14.0%) |
30 (30.0%) | ||
Distribution of
|
Frequency |
Percent | ||
|
E. coli O157:H7 |
Positive |
1 |
1.4% |
|
Negative |
69 |
98.6% | |
|
Total |
70 |
100.0% | |
Association between gender and test results
|
Gender |
Total |
P. value | |||
|
Male |
Female | ||||
|
E. coli O157:H7 |
Positive |
1 (1.4%) |
0 (0.0%) |
1 (1.4%) |
0.335 |
|
Negative |
37 (52.9%) |
45.7 (45.7%) |
69 (98.6%) | ||
Association between age and test results
|
Age |
Total |
P. value | ||||
|
< 20 years |
20 - 40 years |
> 40 years | ||||
|
E. coli O157:H7 |
Positive |
1 (1.4%) |
0 (0.0%) |
0 (0.0%) |
1 (1.4%) |
0.488 |
|
Negative |
28 (40.0%) |
24 (34.3%) |
17 (24.3%) |
69 (98.6%) | ||
Discussion
One of the most commendable aspects of this study is its focus on a region that may be underrepresented in the literature regarding food-borne pathogens. By investigating the presence of O157 in Khartoum, the present study addresses a crucial gap in understanding how this pathogen affects local populations. The choice of using a molecular technique such as PCR for detection is also a notable strength, as it offers higher sensitivity and specificity than traditional culture methods do.
The methodological approach is well structured, beginning with the collection of stool samples from a variety of hospitals. This diversity in sampling can enhance the generalizability of the findings. The use of different microbiological techniques—including culture on MacConkey agar, Gram staining, and biochemical tests—demonstrates a thorough approach to isolate and identify , ensuring that the results are reliable.
Additionally, the study's results highlight a critical concern: while 70% of the samples contained , only 1.4% of the samples tested positive for the more virulent strain O157. This disparity raises important questions about the prevalence of pathogenic strains versus nonpathogenic strains, emphasizing the need for ongoing surveillance and monitoring of food-borne pathogens.
Verocytotoxin-producing O157:H7 emerged as a major food-borne pathogen from the 1980s--1990s, and the disease is often associated with significant mortality in young and elderly patients 26. It causes severe life-threatening hemolytic uremic syndrome (HUS) worldwide 27, 28, 29.
One hundred samples were collected from 54 (54%) males and 46 (46%) females; 70 isolates (70%) were. This result is in agreement with previous reports that revealed that 70% of bacteria are positive for , although this result disagrees with other studies performed for the isolation and detection of O157:H7, which detected (78%) isolates from stool cultures 29, 30.
All isolates in this study were tested via conventional PCR to identify the O157:H7 gene, and the percentage of O157:H7 was 1.4%. This result is in agreement with previous findings that reported O157:H7 (1.14%). This finding was similar to that of other previous studies in which O157:H7 was detected in 1 (0.77%) diarrheic patient. This proportion was lower than that reported in a previous study, which revealed that O157:H7 was detected in 5 isolates (2.3%) that were positive in 210 samples. However, the highest percentage of O157:H7 (6%) was detected29, 30, 31, 32.
Wang ., Getaneh ., Peroutka-Bigus ., and Jenkins . 33, 34, 35, 36 agreed with our results and support our findings.
Future studies including larger sample sizes could increase the statistical power and reliability of the results. Furthermore, a more detailed demographic breakdown of the patients, including their age, health status, and dietary habits, could provide valuable insights into the risk factors associated with O157 infection.
Further details about the PCR process—such as the specific primers used, amplification conditions, and controls employed—would increase the reproducibility of the study. Clearer documentation of these methodological steps would allow other researchers to replicate the study and validate its findings.
Further research including the implications of the findings more thoroughly would benefit from a deeper exploration of these complications and how they may differ among various demographic groups. Providing context about the health impacts of this pathogen, particularly in vulnerable populations such as children and elderly individuals, would strengthen the public health message. Moreover, further studies could address potential environmental and socioeconomic factors contributing to the prevalence of O157 in the region. Understanding the broader context of food safety practices, sanitation, and public health infrastructure in Khartoum would provide a more holistic view of the issue. Finally, the study should consider recommendations for public health interventions on the basis of its findings. Suggestions for improving food safety practices, enhancing community awareness about food-borne illnesses, and implementing regular surveillance programs.
Study limitations
The study's limitations include its focus on patients from Khartoum State, potentially limiting its generalizability across Sudan. Additionally, the sample size is small.
Conclusions
In conclusion, this study contributes to the understanding of O157 in Khartoum State by employing robust molecular techniques to detect this significant pathogen. Its strengths lie in addressing a regional gap in research and utilizing effective identification methods. However, there are several areas for improvement, including the need for a larger sample size, more detailed methodology, and a comprehensive discussion of the findings' implications. Future research can better inform public health strategies aimed at preventing food-borne illnesses and protecting vulnerable populations in the region.
List of abbreviations
CDC: Central Disease Control
EAEC: Enteroaggregative
EHEC: enterohemorrhagic ()
EIEC: Enteroinvasive
EPEC: Enteropathogenic
ETEC: Enterotoxigenic
HUS: Hemolytic uremic syndrome
KIA: Kligler iron agar
PCR: Polymerase chain reaction
SPSS: Statistical Packages for Social Sciences
STEC: Shiga toxin-producing
Declarations
Ethics approval and consent to participate
Ethical approval was obtained from the Ministry of Health Ethical Research Committee in accordance with the Declaration of Helsinki Principles, and the agreement was taken from the Dentistry Hospital Administration before sample and data collection. The patient’s information was highly secure and not used for purposes other than scientific inquiry. The aims and objectives of the study, along with its procedure, methods, risks and benefits, were explained to each participant in easily understandable local language, and written informed consent was obtained from each patient .
Ethical clearance code number
MH-RES/17-022-11
Date: 22/12/2022
Consent for publication
Not applicable.
Availability of data and materials
The data sets used and/or analyzed during the current study are available from the corresponding author upon reasonable request .
Competing interests
The authors declare that they have no competing interests .
Funding
Not applicable.
Authors’ contributions
SAM and SMS conceived the design and carried out the experiments. AAI obtained, analyzed and interpreted the data. RAA, SAM and SMS wrote and revised the manuscript. AAI and RAA provided financial support for all the experiments. All the authors critically reviewed and approved the final draft and are responsible for the content and similarity index of the manuscript .
Acknowledgments
We thank all the participants involved in this research.

